Systems biology meets epigenetics: A computational model explains epigenome dynamics during differentiation
(Phys.org)—Scientists from the Friedrich Miescher Institute for Biomedical Research and the Biozentrum of the University of Basel have published an important proof-of-principle study showing that a computational model can elucidate the interplay of transcription regulators and epigenome dynamics during differentiation. This is critical for a better understanding of the nature of different cell types and disease stages.
Epigenetics and systems biology are two disciplines that created a lot of excitement in the research community in the last decade. It is widely believed that an even bigger potential lies in the combination of both fields, yet experimental and computational scientists have to join forces. Such collaboration has been successfully shaped in the framework of the systemsX.ch project Cell Plasticity. In the realm of epigenetics and systems biology the groups of Dirk Schübeler at the Friedrich Miescher Institute for Biomedical Research (FMI) and of Erik van Nimwegen at the Biozentrum of the University of Basel have accomplished an important proof-of-principle study.
Interplay between chromatin structure and transcription factors
Three years ago, Cell Plasticity set out to elucidate the genome-wide epigenetic and transcriptional events that occur during cell differentiation using a systems biology approach. During differentiation, the process that leads from a pluripotent stem cell to a mature cell, gene expression is adapted continuously. This regulation is coordinated by transcription factors together with dynamic changes in the local organization of chromatin. In the last couple of years, mapping of these chromatin changes such as DNA methylation or histone modifications on a genome-wide scale has become possible. However, it is still unclear how chromatin modifications are targeted to specific loci in the DNA. What is clear is that the regulation of gene expression involves mutual feed-back mechanisms between transcription factors and chromatin structure: chromatin structures hinder or allow the binding of transcription factors on the one hand, and transcription factors can recruit proteins to the DNA that alter chromatin structure on the other hand. The interplay between these two mechanisms then determines the resulting gene expression patterns.
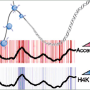
It is this process that was the focus of the study of Schübeler and van Nimwegen, which was published recently in Genome Research. Since there are between 1'500 and 2'000 transcription factors, the contribution of each to chromatin structure during differentiation cannot be tested experimentally. The scientists thus aimed to model the relationship between computationally predicted transcription factor binding sites and genome-wide changes in chromatin structure. In more general terms, they wanted to see if their computer model allows them to identify the transcription factors that lead to changes in the chromatin structure relevant for differentiation.
Exemplary collaboration
Indeed, in an exemplary collaboration where the van Nimwegen PhD student Phil Arnold developed the computational model and Anne Schöler, graduate student in Schübeler's team, validated the results experimentally, the scientists identified several sequence motifs that regulate the dynamics of specific chromatin marks during neuronal differentiation, namely the motifs bound by REST and the SNAIL family of transcription factors.
"These results are important for our systemsX.ch project because they prove that a systems approach to elucidate the genome-wide epigenetic and transcriptional events during differentiation is actually fruitful," comments Schübeler. And van Nimwegen complements that "This work is a perfect example of how a tight integration of computational work with follow-up experiments can lead to insights far beyond what these approaches could have accomplished by themselves."
The same approach is applied to other models of differentiation and disease within the Cell Plasticity network, which is headed by Susan Gasser, director of FMI, and includes colleagues at the FMI, the Biozentrum, the Department of Biomedicine, the University Hospital and Novartis, making Basel a hotspot for systems biological approaches in epigenetics.
More information: Arnold P, Schöler A, Pachkov M, Balwierz P, Jørgensen H, Stadler MB, van Nimwegen E, Schübeler D (2012) Modeling of epigenome dynamics identifies transcription factors that mediate Polycomb targeting, Genome Res. 2012 Sep
www.cellplasticity.org/
www.systemsx.ch/
Journal information: Genome Research