Structure discovered for promising tuberculosis drug target
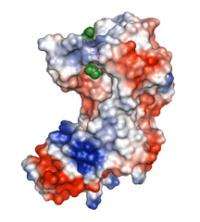
Researchers at Johns Hopkins have figured out the three-dimensional shape of the protein responsible for creating unique bonds within the cell wall of the bacteria that cause tuberculosis. The bonds make the bacteria resistant to currently available drug therapies, contributing to the alarming rise of these super-bacteria throughout the world.
With the protein structure in hand, the scientists say, drug designers have a clear way forward for weakening the cell wall and killing these deadly bacteria. Their results are reported in a paper published online Oct. 25 in Structure.
"We've known for a while that this protein would make a good drug target, but without a structural model, drug discovery is like blindly choosing random objects to fit into a small hole of unknown shape and size. The results of our study have removed the blindfold," says L. Mario Amzel, Ph.D., professor and director of the Department of Biophysics and Biophysical Chemistry at The Johns Hopkins University School of Medicine.
The Johns Hopkins team used a technique called X-ray crystallography to scatter radiation off a specially prepared portion of the enzyme that forms the unique molecular bonds within the cell wall of Mycobacterium tuberculosis. They then used information about the direction and intensity of the radiation scattered to build a 3-D model of the arrangement of atoms in the enzyme.
Mario A. Bianchet, Ph.D., assistant professor of neurology at Johns Hopkins and a member of the research team, says the challenge of TB infection is that most of the long and costly standard drug treatment is just to get rid of the roughly 1 percent of bacteria that persist after the first week of a patient's treatment. "The 'persisters' resist in part because of unique bonds within their cell walls. Their cell walls form a thick, three-layered boundary between the bacteria and the outside world, including a middle layer of interlocking molecules, called peptidoglycans, that form a network resembling a chain-link fence," says Bianchet.
Peptidoglycans are long chains of individual sugar molecules with short protein branches extending from every other sugar on alternating sides of the chain. Specific enzymes bond the protein branches to each other to create a meshwork. In most species of bacteria, Amzel says, the majority of the bonds between these branches are created between position 4 on one branch and position 3 on an opposing branch. In M. tuberculosis, however, the majority of the bonds are created between positions 3 on both branches. The most common antibiotics interfere with the enzyme that creates the 4-3 bonds, which is enough to destabilize the cell wall and kill most of the TB bacteria.
The bacteria that persist have a particularly high level of 3-3 bonds between the peptidoglycan chains in their cell walls. These 3-3 bonds are created by a different enzyme, the one that Amzel and Bianchet studied, which is not specifically targeted by any current drugs. In addition to showing the structure of the enzyme, the team also showed a peptidoglycan molecule inside the action site where the 3-3 bonds are made, giving drug designers even more details about the way the protein works.
Amzel adds that "beyond fighting TB, the structure of this enzyme may help us fight other disease-causing bacteria that have similar enzymes, such as Enterococcus faecium and the spore-forming, drug-resistant Clostridium difficile."
More information: Structure doi: 10.1016/j.str.2012.09.016
Journal information: Structure
Provided by Johns Hopkins University School of Medicine
















