September 14, 2012 feature
From vitro to vivo: Fully automated design of synthetic RNA circuits in living cells
(Phys.org)—Synthetic biology combines science and engineering in the pursuit of two general goals: to design and construct new biological parts, devices, and systems not found in nature; and redesign existing, natural biological systems for useful purposes. For synthetic biologists a key goal is to use RNA to automatically engineer synthetic sequences that encode functional RNA sequences in living cells. While earlier RNA design attempts have mostly been developed in vitro or needed fragments of natural sequences to be viable, scientists at Institut de biologie systémique et synthétique in France have recently developed a fully automated design methodology and experimental validation of synthetic RNA interaction circuits working in a cellular environment. Their results demonstrate that engineering interacting RNAs with allosteric behavior in living cells can be accomplished using a first-principles computation.
Drs. Alfonso Jaramillo, Guillermo Rodrigo, and Thomas E. Landrain had to address several challenges in their study. "It is common practice – and unavoidable – to use computational algorithms to aid in the design of RNA molecules," Jaramillo tells Phys.org. For example, he illustrates, computing minimum energy conformation, since one single nucleotide can stabilize an alternative conformation. Until now researchers have used computer assisted design to design synthetic RNAs that could combine functional fragments from known RNAs.
"The combination of known fragments reduced the number of possible nucleotide sequences and would enable a human-driven iteration in reasonable time," Jaramillo explains. This approach required a lot of significant human effort to design a single sequence, where the designer would combine the fragments with some educated guesses and random nucleotides to fill the gaps. At that point, other computational tools would be used to verify each possible prototype and improve it. "We wanted to go further, so we asked whether we could teach the computer to do all this design iterations automatically, such that it could suggest a solution that could be synthesized and transformed in living cells," Jaramillo adds.
Jaramillo comments that if the computer performs all design iterations unsupervised, it can explore a larger search space, freeing researchers from having to use known sequence fragments, and thus attempt a full sequence design (where all of the nucleotides come from random suggestions followed by computer selection). "To instruct the computer to do this, we incorporated what we thought were the essential physical properties of interacting RNAs in living cells by employing the evolutionary computation techniques used in artificial intelligence to develop software that could code without human supervision. The computer could then explore millions of prototype sequences instead of a few."
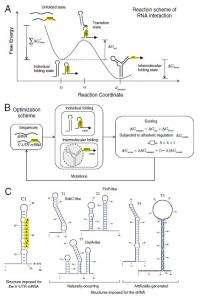
This evolutionary computation technique relies on mimicking the relevant steps of natural evolution, that is, the iterative improvement of a given solution by using selection. "However, we don't have to be slaves of analogy and are free to consider what we think is more relevant to our problem," Jaramillo points out. "We would start from a random sequence and would randomly modify it by applying simulated annealing techniques, implemented by a Metropolis Monte Carlo algorithm," which solves a problem by generating suitable random numbers and observing that fraction of the numbers obeying some property or properties. "Contrary to natural evolution, our walks would not be completely adaptive but we could allow a decrease in fitness. We aim at the engineering of an ensemble of RNA species that could interact in a predefined way. Our first challenge was that in living cells, such molecules are very prone to degradation if they do not have a stable structure."
The scientists therefore imposed that all our RNA species would have such structure – but this produced another challenge. "Unlike unstructured RNAs, such structure prevents RNA molecules from mutually interacting. We approached this by including a nucleation site, allowing an initial intermolecular contact that could disassemble the folded structure of the interacting partners." This nucleation site consisted of a small fragment of an unstructured sequence. "This stabilization could be seen in energetic terms as the lowering of the activation free energy barrier, where we lower the energy of a high energy intermediate state."
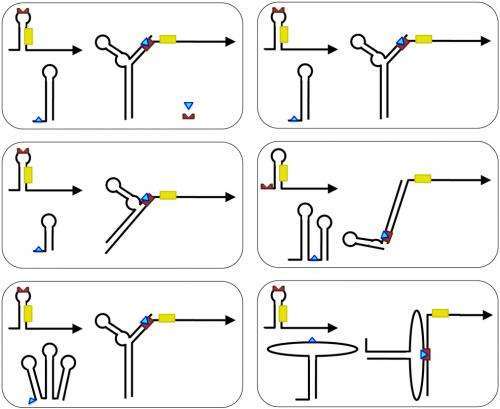
As they relied on structured RNAs that would undergo post-interaction conformational changes, the scientists tested it by exploring a challenging problem involving allosteric RNA p and so chose to work with Escherichia coli. "We selected the problem of designing positive riboregulators because, due to design challenges, only a handful of them have ever been engineered by humans. Such positive riboregulators are RNA that, once expressed, could activate translation machinery." This problem was not only challenging, but also useful for biotechnology, where there are very few molecules genetically encoded able to activate gene expression (contrary to the relative ease of engineering gene expression repressors). "As our methodology relied on fixing the secondary structure of the interacting RNAs, we tested several possibilities as well as alternative interaction mechanisms where the initial hybridization could be done in different ways."
After publishing the PNAS manuscript, Jaramillo adds, the team further validated the orthogonality (the ability to selectively translate mRNA) of their RNAs in E. coli. They're also constructing a XOR gate device working inside the cell – something never done in bacteria and just recently achieved in mammals1.
The researchers are planning to extend the methodology to include the RNA-small molecule interactions and the incorporation of known functional RNA sequence fragments (such as ribozyme sequences) to create complex RNA interactions never seen before. "We've already succeeded in experimentally validating in E. coli a new type of such an interaction, consisting of an inactivated riboregulator that could be activated by a ribozyme after the introduction of a small-molecule inducer."In this type of reaction, the number of different species is not conserved, as after the introduction of the inducer we get a RNA cleavage. "We've named this new riboregulator-ribozyme chimera a regazyme, and have also validated the full design of a riboswitch.
Jaramillo also notes that other research might benefit from their findings, including the high-throughput design of new regulators for large-scale engineering projects. "Also, we can foresee using allosteric RNAs to sense mRNAs by being subject to a conformational change after binding that could trigger a reporter." This would open the way to genetically-encoded and non invasive monitoring of gene expression dynamics – an important and unmet challenge in biophysics. "We're also exploring the use of RNA," Jaramillo concludes, "to create artificial signal transduction cascades."
More information: De novo automated design of small RNA circuits for engineering synthetic riboregulation in living cells, PNAS September 4, 2012, doi:10.1073/pnas.1203831109
1Related: Programmable single-cell mammalian biocomputers, Nature 487, 123–127 (05 July 2012), doi:10.1038/nature11149
Journal information: Proceedings of the National Academy of Sciences , Nature
Copyright 2012 Phys.org
All rights reserved. This material may not be published, broadcast, rewritten or redistributed in whole or part without the express written permission of PhysOrg.com.