Talin links cytoskeleton and cell membrane in migrating and dividing cells, researchers find
Cells can show a remarkable range of motility, creeping over substrates using a variety of pushes, pulls, stretches, and drags to get from A to B. This range of motion is achieved through the concerted efforts of motor proteins and structural complexes collectively known as the cytoskeleton. In addition to propulsion, however, cells also need to find footholds on surfaces in order to get the traction needed to advance or withdraw. Mobile single-celled organisms, such as the amoeba Dictyostelium, provide excellent living models for studying the molecular basis of such mechanisms, as they spend much of their lives solitary and on the crawl.
New work by Masatsune Tsujioka and colleagues in the Electron Microscope Laboratory reveals how a protein called talin A links the cytoskeletal complex actomyosin to the plasma membrane of Dictyostelium cells as they move and divide. Published in the Proceedings of the National Academy of Sciences, this work was conducted in collaboration with colleagues at Yamaguchi and Kyoto Universities, the University of Edinburgh, and the RIKEN Quantitative Biology Center (QBiC).
Cortical actomyosin is a contractile fiber-like complex of filamentous actin and the motor protein myosin II, and is the driving force behind many forms of changes in cell morphology and movement. This activity requires connections between this cytoskeletal molecular complex and the membrane of the cell, but how this is accomplished has remained something of a mystery. Talin, a protein known to function in integrin-mediated cell-substrate adhesions, has been implicated as a candidate for this role, as loss of talin function has been shown in previous studies to affect bond between cells and their substrate. Knowing this, Tsujioka opted to study the question more closely in Dictyostelium as it possesses only a single myosin II gene, simplifying the task of studying it in isolation, and it shows a remarkable range of motile behaviors.
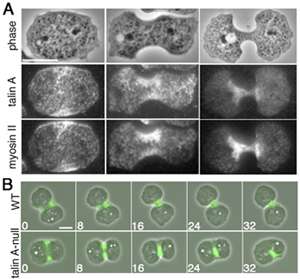
He began by examining the subcellular localization of talin A (the Dictyostelium homolog of mammalian talins). Using a GFP-tagged construct and immunostaining techniques, Tsujioka found that this protein co-localized with myosin II at the back end of migrating amoebae. He next used a sophisticated piece of kit called a total internal reflection fluorescence microscope, which enables very thin sections of a sample to be visualized, to check for direct binding between the two, but was intrigued to find that while talin A and myosin II remained close to one another, they were not directly connected. Both did co-precipitate with actin, raising the possibility that this fibrous protein served as the link.
When Tsujioka added talin A to Dictyostelium cells lacking both myosin II and endogenous talin, he found that it spread throughout the entirety of polarized migrating cells, rather than just in the rear, indicating that its distribution relies on myosin. To test whether the talin A protein plays a functional role in cell motility, the team set up a system for observing Dictyostelium cells as they moved, taking advantage of this amoeba's well-known taste for cAMP. When talin A-null cells in a microcapillary were presented with a chemical gradient, they would set off to follow it, but their posterior ends often remained stuck behind them – resembling a long tail that failed to detach from the substrate. Suspecting that this sticky predicament might be mediated by a cell-substrate adhesion molecule called SibA, they next generated double knockouts for both proteins and found that the cellular tails vanished.
Actomyosin also plays a role in mitotic cell division, so Tsujioka next asked whether talin might be at work in this process as well. As in migrating cells, talin co-localized with myosin II, in this case mid-cell near the cleavage furrow, and again talin's accumulation appeared to rely on myosin II. In dividing wildtype cells, this furrow gradually folds inward, a form of movement called ingression, eventually splitting the cell into a pair of daughter cells. In cells in which talin A was deleted, however, the furrow was wider and longer than in wildtype, and responded differently to mechanical stresses (in this case, being sandwiched tightly under a sheet of agarose), which appeared to be due to a disconnect between the actomyosin contractile ring and the plasma membrane.
As a last step, the lab conducted a structural analysis of the talin A protein to look for possible binding sites that might account for its apparent coupling role. They found that while the talin C-terminal contains an actin binding site, explaining its bond with actomyosin, a locus on its N-terminal domain bound with a phospholipid called PtdIns(4,5)P2, which is also enriched in the tail ends of migrating cells, suggesting that this might be protein that activates talin A as a conveyor of contractile force in migrating and dividing cells.
"The fact that a similar separation between the plasma membrane and the underlying actin has been seen to occur in talin mutant cells in mammals as well suggests that that this discovery of a novel function for the molecular in Dictyostelium may be evolutionarily conserved," says Tsujioka. "We hope to dig deeper in future analyses of this new function and to identify other molecules involved in constructing and regulating these links."
More information: www.pnas.org/content/109/32/12992.long
Journal information: Proceedings of the National Academy of Sciences
Provided by RIKEN