This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Novel software tool enables quality control independent of omics molecular types and can be used on multiple platforms

Quality control (QC) strategies for monitoring and assessing instrument performance during mass spectrometry analysis is crucial for ensuring that the data generated is high-quality, reproducible, and accurate.
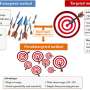
Particularly for metabolomics and lipidomics, traditional QC approaches often involve manual inspection of the data, which is time-consuming, subjective, prone to human error, and not viable for high-throughput studies with hundreds of samples.
A team of scientists from Pacific Northwest National Laboratory (PNNL) and the Environmental Molecular Sciences Laboratory (EMSL) have developed PeakQC, a new software tool for automated quality control of mass spectrometry data.
The findings are published in the Journal of the American Society for Mass Spectrometry.
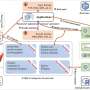
Unlike existing QC tools that focus on specific types of molecules, PeakQC can analyze data from various "omics" fields, like proteomics, metabolomics, and lipidomics. The software performs global and detailed QC assessments and works with different instruments and experimental setups, including liquid chromatography and ion mobility spectrometry.
By providing automated QC that works across different types of molecules and experimental setups, this new software can help researchers maintain data reliability and catch problems early. This could lead to more efficient use of resources, better reproducibility in experiments, and ultimately more trustworthy scientific conclusions.
The tool's flexibility makes it particularly valuable as more studies combine multiple "omics" approaches. PeakQC enables the collection of high-quality data in large-scale user projects at EMSL and within PNNL. The tool not only enables comparison of outputs from multiple mass spectrometry approaches, but also facilitates the collection of reproducible data.
The software program uses advanced algorithms and machine learning to analyze data from various instrument types and experimental methods, including both data-dependent and data-independent acquisition modes.
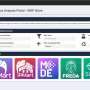
The software is a desktop and stand-alone tool that generates diagnostic plots and metrics without relying on other molecular identification tools, which are often complicated to set up and use. It is freely available, easy to use, and requires no installation. Once downloaded, users can launch the tool on their own device.
PeakQC can use either user-specified ions or automatically detected ions to extract quality control metrics. This approach allows it to pinpoint specific causes of performance issues, unlike some existing tools that only provide general quality assessments.
By offering a unified approach to quality control across different "omics" fields, PeakQC represents a significant advancement in mass spectrometry data analysis. Its development highlights the growing importance of robust, automated quality control in large-scale scientific studies using mass spectrometry.
More information: Andrea Harrison et al, PeakQC: A Software Tool for Omics-Agnostic Automated Quality Control of Mass Spectrometry Data, Journal of the American Society for Mass Spectrometry (2024). DOI: 10.1021/jasms.4c00146
Provided by Environmental Molecular Sciences Laboratory