This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Simplifying complexities in bioinformatics: A desktop suite for multi-omics data analysis and visualization
The revolution in high-throughput sequencing and mass spectrometry technologies, such as Illumina, PacBio, and 10X Genomics, have enabled the production of vast amounts of biological data. However, the intricate data formats and the need for specialized analytical pipelines pose significant challenges, especially for experimental biologists who may find command-line interfaces, coding, and scripting daunting.
In September 2023, Horticulture Research published research titled "OmicsSuite: a customized and pipelined suite for analysis and visualization of multi-omics big data."
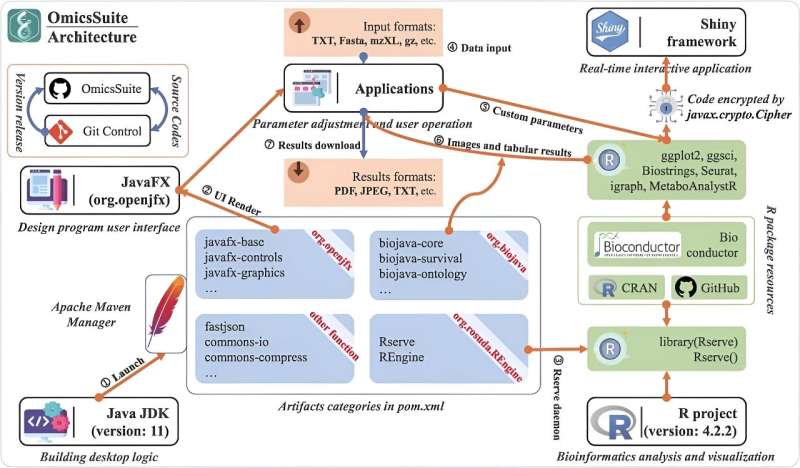
OmicsSuite is a comprehensive desktop suite that facilitates multi-omics data analysis and visualization, integrating 175 sub-applications across various analysis tasks. Developed with JavaFX for user interaction and R for computational analysis, it supports a wide range of multi-omics studies, including plant chloroplast genome analysis, GWAS, transcriptomics, metabolomics, and single-cell RNA-Seq.
The key features of OmicsSuite include user-friendly interfaces(UI), comprehensive coverage, highly readable Multi-omics raw data, and output of results through intuitive UI elements.
The OmicsSuite utilizes BioJava for bioinformatics functions and over 300 open-source packages for diverse analytical capabilities. It excels in integrating visualization tools such as ggplot2 and Shiny applications, offering a holistic analysis environment without requiring extensive setup or computational resources.
Its architecture enables seamless interaction with the R environment for real-time data processing, supporting vast multi-omics data types from LC-MS data to single-cell genomics formats. OmicsSuite's UI design enhances user experience with a modern layout, easy navigation, and comprehensive tutorial support; a computer device with a 4-core CPU, 4G memory, and 256G storage can perform normal operations.
Overall, this toolset simplifies the complex data analysis workflow, making it accessible to researchers in plant breeding and molecular mechanism studies, evidenced by its growing adoption and positive community feedback.
More information: Ben-Ben Miao et al, OmicsSuite: a customized and pipelined suite for analysis and visualization of multi-omics big data, Horticulture Research (2023). DOI: 10.1093/hr/uhad195
Provided by NanJing Agricultural University