This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Computer model boosts detection of cell-to-cell communication

A computer model developed by UT Southwestern Medical Center researchers significantly enhances the ability of scientists to detect communication between cells, according to a new study published in Nature Methods.
The model, called Spacia, could help advance understanding of a wide range of diseases including cancers, autoimmune disorders, infectious diseases, and developmental abnormalities.
"Cell-to-cell communication (CCC) is incredibly important for all life forms. With Spacia, we are able to decipher it better than ever before," said Tao Wang, Ph.D., Associate Professor in the Peter O'Donnell Jr. School of Public Health and in the Center for the Genetics of Host Defense at UT Southwestern. He is a member of the Population Science & Cancer Control Research Program in the Simmons Cancer Center.
Dr. Wang co-led the study with Yang Xie, Ph.D., Professor in the O'Donnell School of Public Health and the Lyda Hill Department of Bioinformatics and Associate Dean for Data Sciences at UT Southwestern Medical School. Drs. Wang and Xie are investigators in the Quantitative Biomedical Research Center in the O'Donnell School of Public Health.
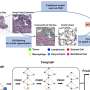
CCC is necessary for a vast range of biological processes, including development, health maintenance, and disease progression. In recent years, researchers have developed experimental techniques that provide information on the activity of genes in individual cells (single cell sequencing), and even the location of cells (spatially resolved transcriptomics, SRTs), which yield critical information for inferring CCCs.
However, programs that analyze the wealth of data generated by these techniques to extract accurate CCC relationships have several drawbacks. For example, some programs take an average reading of gene activity among groups of spatially neighboring cells, losing single-cell resolution, and others are only able to detect communications between cells in known regulatory pathways.
To overcome these issues, Drs. Wang and Xie and their colleagues used a mathematical technique called multi-instance learning (MIL) to develop Spacia for inferring CCCs from SRT data. MIL, a subset of machine learning, is well known in the computer science field; however, it has rarely been explored for biomedical use, Dr. Xie explained.
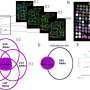
The researchers tested Spacia in a variety of contexts using data generated by SRTs, gathering significant insights. For example, using an SRT dataset from prostate cancer tissue, Spacia found that several cell types in the tumor microenvironment participate in a phenomenon called epithelial-mesenchymal transition, an important contributor to metastasis.
Deploying Spacia on a pan-cancer SRT dataset that included breast, colon, skin, and lung cancers, among other types, the researchers found that B cells, a type of immune cell, react to signaling from tumor cells targeted by immunotherapy drugs known as checkpoint inhibitors.
They also discovered a different CCC signature that accurately predicted cancer patient survival rates and their response to checkpoint inhibitors.
"Our study highlights the power of integrating spatial and transcriptomic data to uncover hidden cellular interactions that drive disease progression and treatment resistance," said Dr. Xie. "Spacia represents a significant advancement in our ability to translate molecular insights into clinical applications, ultimately improving patient care."
Dr. Xie noted that Spacia is presently most useful for biological research. But eventually, as the current high cost of SRT declines, doctors may use this tool to identify drug targets specific to individual patients' diseases, dramatically improving personalized medicine.
More information: James Zhu et al, Mapping cellular interactions from spatially resolved transcriptomics data, Nature Methods (2024). DOI: 10.1038/s41592-024-02408-1
Journal information: Nature Methods
Provided by UT Southwestern Medical Center