This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Scientists reveal mechanism of maternal protein Pramel15 in promoting DNA demethylation in mouse zygotes
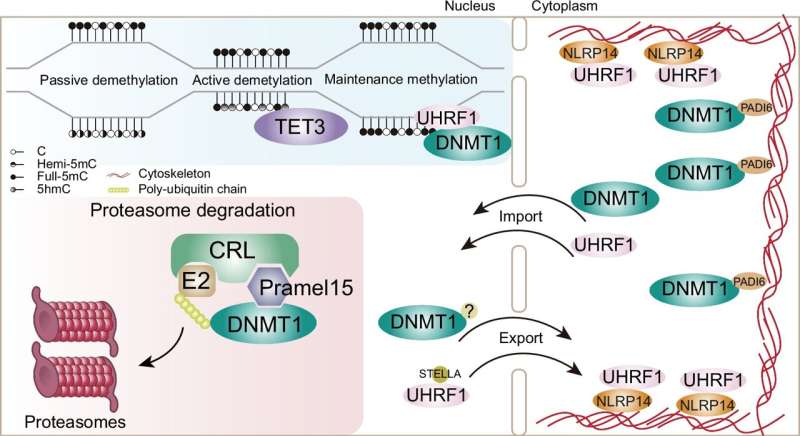
DNA methyltransferase 1 (DNMT1) is an important DNA methyltransferase that maintains DNA methylation. The retention of DNMT1 and its cofactor UHRF1 in the cytoplasm is considered a major reason for passive DNA demethylation in the oocyte and early embryonic development.
However, the mechanisms by which DNMT1, UHRF1, and other proteins are regulated and how they play appropriate roles in the DNA methylation reprogramming process have not yet been elucidated.
A new study from the research group led by Prof. Zhu Bing at the Institute of Biophysics of the Chinese Academy of Sciences found that the maternal factor Pramel15 in mice can mediate DNMT1 degradation in the zygotic nucleus, fine-tuning nuclear DNMT1 protein abundance.
This study reveals, for the first time, the presence of a proteasome-dependent regulatory mechanism for DNMT1 in the early embryonic nucleus, which aids in the DNA demethylation of the zygote after fertilization. The research is published in the journal Nature Communications.
Researchers identified a novel DNA methylation regulatory gene, Pramel15, from the cDNA library of mouse oocytes in their previous study. Overexpression of Pramel15 in somatic cells interferes with the maintenance of DNA methylation during DNA replication by degrading DNMT1.
Further studies in HEK293 and mouse embryonic stem cells revealed that Pramel15 acts as a substrate recognition subunit in the Cullin5 E3 ubiquitin ligase complex, promoting the degradation of DNMT1 through the ubiquitin-proteasome pathway.
By constructing Pramel15 knockout mice, the researchers found that there exists a major DNMT1 proteasomal degradation pathway mediated by Pramel15 in fertilized eggs. Whole-genome DNA methylation sequencing (WGBS) of oocytes, fertilized eggs, and two-cell embryos showed that the absence of Pramel15 leads to a random increase in DNA methylation across the genome.
This indicates that, during early embryonic development, in addition to transporting DNMT1 and UHRF1 and restricting them to the cytoplasm, there is also a Pramel15-mediated regulatory mechanism controlling the levels of DNMT1 in the nucleus, which modulates the efficiency of DNA methylation reprogramming.
The research illustrates that Pramel15 can regulate the levels of DNMT1 in the nucleus during DNA replication in fertilized eggs, thereby aiding in DNA methylation reprogramming in early embryos.
This finding reflects the complexity of the regulatory mechanisms of DNA methylation reprogramming in early embryos and provides insights for improving the efficiency of induced cell reprogramming.
More information: Jiajun Tan et al, Pramel15 facilitates zygotic nuclear DNMT1 degradation and DNA demethylation, Nature Communications (2024). DOI: 10.1038/s41467-024-51614-0
Journal information: Nature Communications
Provided by Chinese Academy of Sciences