This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
International team discovers key protein that helps cells maintain their identity
A discovery regarding Mrc1 (Mediator of Replication Checkpoint 1)—a fission yeast protein involved in DNA replication—has been published in Cell. The discovery is the result of an international research collaboration, led by Professors Genevieve Thon and Anja Groth at the University of Copenhagen.
Dr. Sebastian Charlton, shared first author of the work, had been researching Mrc1 for many years at the Thon laboratory at the Department of Biology. "We knew this protein was important for maintaining the heterochromatic state in cells. We had a good idea of how it worked, but while we had experimental data, we didn't have the tools in our lab to confirm it on a molecular level," he explains.
To test their hypothesis by advanced genomics developed by the Groth laboratory, Dr. Charlton joined forces with Dr. Valentin Flury, shared first author from the Groth group at the Novo Nordisk Foundation Center for Protein Research (CPR).
"But our collaboration didn't stop there," he explains. "We also worked with structural biologists at CPR and with researchers at the Tokyo Metropolitan Institute of Medical Science and the Hubrecht Institute in Utrecht. This successful collaboration not only proved our initial hypothesis, but also drove further experiments which revealed the surprising duality in Mrc1 function."
The role of Mrc1 in DNA replication
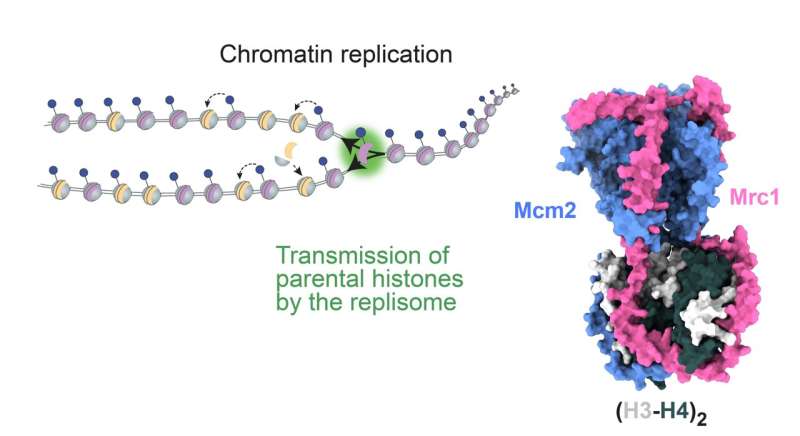
The fork protection complex (FPC) is a group of proteins, including Mrc1 (Claspin in humans), that play a crucial role in replication of DNA. The new discovery reveals that FPC also recycles parental histones, with their specific epigenetic marks, onto the two newly synthesized DNA strands. This process is crucial for maintaining epigenetic memory, as it allows the daughter cells to inherit the same epigenetic landscape as the parent cell.
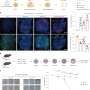
Dr. Charlton and Flury's experiments showed that, during DNA replication, Mrc1 operates as a central coordinator of parental histone inheritance by binding the tetramer formed from histones H3 and H4.
Their research went on to show that mutations in the key connector domain of Mrc1 disrupt the proper distribution of parental histones to the lagging strand during DNA replication. This effect is comparable to disruptions caused by mutations in the histone-binding region of Mcm2—another protein already known to be involved in histone recycling during DNA replication.
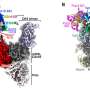
The investigations included structural predictions by AlphaFold, validated by biochemical and functional analysis, which suggest that Mrc1 and Mcm2 collaboratively bind H3-H4 tetramers, with the Mrc1 connector domain playing a key role in linking histones to Mcm2. Thus, Mrc1 and Mcm2 form a co-chaperone complex, ensuring distribution of histones to the lagging strand during DNA replication.
In mutants of Mrc1, the researchers found that the gene silencing mediated by H3K9me heterochromatin is compromised and, amazingly, the de-silenced active state was inherited asymmetrically similar to parental histones. This finding underscores the importance of histones in carrying and transmitting epigenetic information, which maintains stable silencing of genes.
Efficient recycling to both daughter strands
Further experimental analyses revealed that in fact Mrc1 juggles histones in multiple ways: the connector domain directs histones to the lagging strand as described above, while another histone-binding region is required for recycling histones onto the leading strand.
"Previously a role of Mrc1 in transmission of parental histones was not known. Now we have shown that Mrc1 is required for efficient histone recycling to both daughter strands," says Dr. Charlton.
"It appears that Mrc1 toggles histones between both the lagging and leading DNA strands, in part by intra-replisome co-chaperoning with Mcm2. This ensures both daughter cells inherit the correct epigenetic marks, which is essential for preserving gene expression patterns during cell division."
Implications for future research
Maintaining chromatin landscapes across many cell generations is vital for developing and maintaining the multiple cell types in multi-cellular organisms. Losing cellular identity is an underlying cause of many diseases such as cancer, and of aging, where evidence suggests that the chromatin landscape deteriorates over time. Dr. Flury explains that the research team had a "Eureka moment" when they mutated the histone binding sites in the mammalian homolog Claspin and observed a defect in parental histone transmission as with Mrc1 mutants in fission yeast cells.
"I don't think we can estimate the full potential of our discovery yet, but we have revealed a very fundamental mechanism that maintains cell identity which, if it can be manipulated, could have significant implications for future medical research," Dr. Flury concludes.
More information: Sebastian Jespersen Charlton et al, The fork protection complex promotes parental histone recycling and epigenetic memory, Cell (2024). DOI: 10.1016/j.cell.2024.07.017
Journal information: Cell
Provided by University of Copenhagen