August 19, 2024 report
This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Biochemists engineer proteins that can transition between assembly and disassembly via allosteric control
A team of biochemists at the University of Washington has developed a means for engineering proteins that can transition between assembly and disassembly via allosteric control. In their paper published in the journal Nature, the group details their engineering process and how well it has worked thus far during testing.
A. Joshua Wand, with Texas A&M University, has published a News and Views piece in the same journal issue explaining why being able to get a protein to assemble or disassemble in the presence of an effector has been an important goal of chemists and outlines the work done by the team on this new effort.
Prior research has shown that if chemists could build proteins that assemble themselves into desired shapes on command, the results could be used for highly specific purposes, such as building a cage-shaped protein to carry a drug to a certain part of the body for therapeutic purposes.
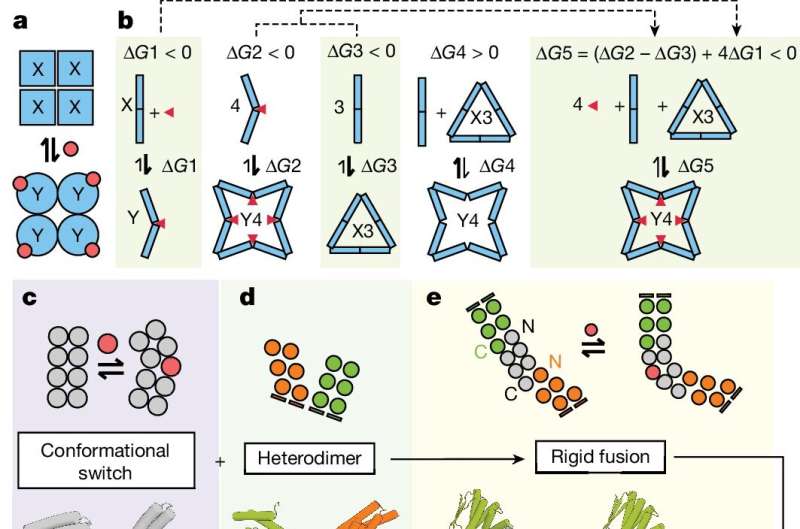
Prior research has also shown that if such a mechanism could be developed, via a process known as allosteric regulation, a specific trigger would be needed—one that chemists refer to as an effector. To achieve this feat, the research team used several techniques they previously developed.
One such technique involved using an AI app to help predict a protein structure given a list of attributes. Another involved creating a protein with a hinge that was able to take on two different forms. And a third involved a way to connect certain proteins together.
The new technique involved using several of these techniques together. For example, they used an AI app to design a desired protein, one that had dual ridged arms and a hinge—such a protein, the team noted, would be stable yet could bend at its hinge. They also noted that the hinge could be used as a sensor to detect the effector.
They then used the AI design to build a real-world protein using their hinge-like proteins and their newly developed technique for connecting them—and the sensor on the hinge to respond to a given effector.
To test their building-block ideas, the research team created several proteins with different shapes using different effectors. One such example was a protein that took on a ringlike structure when it came into contact with a peptide-based effector. The team also found they could create proteins that would grab on to other proteins they built, resulting in new structures.
More information: Arvind Pillai et al, De novo design of allosterically switchable protein assemblies, Nature (2024). DOI: 10.1038/s41586-024-07813-2
A. Joshua Wand, How to design a protein that can be switched on and off, Nature (2024). DOI: 10.1038/d41586-024-02242-7
Journal information: Nature
© 2024 Science X Network