This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Unraveling the DNA mystique of Saposhnikovia divaricata: New horizons in herbal medicine
Scientists have decoded the genetic blueprint of Saposhnikovia divaricata, a traditional medicinal herb. Their research provides a detailed genome sequence, shedding light on the plant's evolutionary adaptations and the genetic foundations of its therapeutic benefits. Identifying crucial genetic markers, the study paves the way for enhancing the medicinal efficacy of this age-old herbal remedy through targeted genomic breeding strategies.
Despite its revered status in traditional Chinese medicine, Saposhnikovia divaricata has not been thoroughly studied genetically. This herb, used to treat rheumatism and skin conditions, holds untapped potential due to unknown genetic and metabolic profiles. The recent study aims to fill this gap by mapping its genome, offering insights that could improve its medicinal properties through advanced breeding and biotechnological methods, potentially leading to superior therapeutic applications.
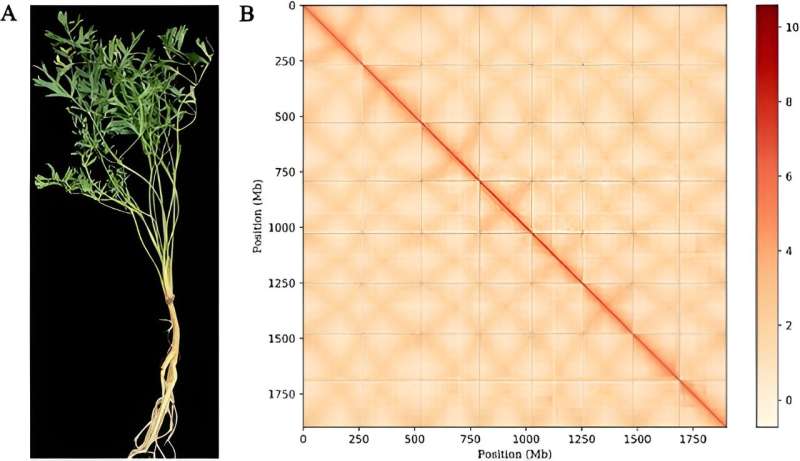
Published in April 2024 in Horticulture Research, this collaborative research involved Jilin Agricultural University and various other institutions, including international partners from the University of British Columbia. Leveraging cutting-edge sequencing technologies, the study delivers a chromosome-level genome assembly, providing a deeper understanding of the plant's intricate genetic structure and its adaptation strategies.
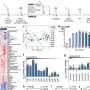
The genomic analysis of Saposhnikovia divaricata uncovered a genome size of 2.07 Gb, rich in repetitive sequences and marked by a notable whole-genome duplication. These characteristics indicate a vast genetic diversity and adaptability, essential for its survival and medicinal effectiveness.
The study pinpointed several gene families crucial for synthesizing key pharmacological agents like flavonoids and plant hormones. Furthermore, comparative genomic studies with related species illuminate unique evolutionary paths and gene expansions that enhance its distinct pharmacological properties, crucial for future efforts to boost the plant's medicinal value.
Dr. Zhong-Ming Han, one of the study's senior authors, stated, "This genome assembly opens new paths for the biotechnological use of Saposhnikovia divaricata. Understanding its genetic and metabolic pathways is foundational for developing enhanced varieties with greater therapeutic potential."
The research holds significant implications beyond traditional applications, opening doors to pharmaceutical innovations. With an improved understanding of the genetic elements driving its pharmacological effects, there is potential for creating more effective medicinal derivatives from Saposhnikovia divaricata, merging ancient remedies with modern medical technology.
More information: Zhen-Hui Wang et al, Genomic, transcriptomic, and metabolomic analyses provide insights into the evolution and development of a medicinal plant Saposhnikovia divaricata (Apiaceae), Horticulture Research (2024). DOI: 10.1093/hr/uhae105
Journal information: Horticulture Research
Provided by NanJing Agricultural University