This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Researchers develop a tool for visualizing single-cell data
Modern cutting-edge research generates enormous amounts of data, presenting scientists with the challenge of visualizing and analyzing it. Researchers at the Helmholtz Institute for RNA-based Infection Research (HIRI) in Würzburg and the Technical University of Applied Sciences Würzburg-Schweinfurt (THWS) have developed a tool for visualizing large data sets.
The sCIRCLE tool allows users to explore single-cell analysis data in an interactive and user-friendly way. Their results have been published in the journal NAR Genomics and Bioinformatics.
Antibiotic resistance is rising worldwide and poses a major challenge to health care systems. Understanding the underlying processes of how and why some bacteria develop such defenses is critical.
Differences in bacterial gene expression—in which the information in a gene is read—could provide clues.
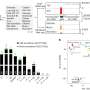
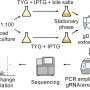
Bacterial single-cell RNA sequencing, or "scRNA-seq," has become an essential tool for analyzing these variations. This technology provides a detailed picture of gene expression in a single cell at a specific point in time. However, the method generates huge amounts of data that are difficult for researchers to visualize and analyze.
Accordingly, there is a great need for new approaches and tools to display and understand this information.
Researchers at the Helmholtz Institute for RNA-based Infection Research (HIRI) in Würzburg, a site of the Helmholtz Center for Infection Research (HZI) in Braunschweig in cooperation with the Julius-Maximilians-Universität Würzburg (JMU), and designers at the Technical University of Applied Sciences Würzburg-Schweinfurt (THWS) have now developed such a tool.
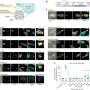
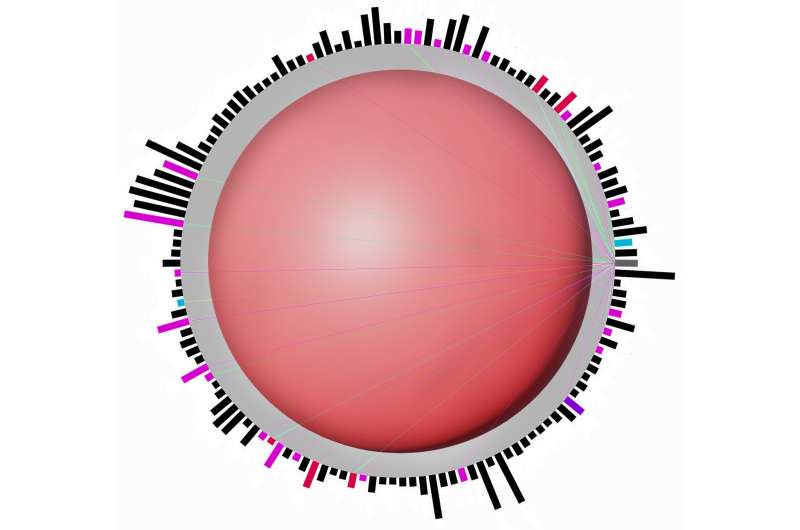
Their desktop application, called sCIRCLE (single-Cell Interactive Real-time Computer visualization for Low-dimensional Exploration), enables interactive 3D visualization of scRNA-seq data. It allows users to view single cells, enriched with metadata, from different angles and in real-time.
A variety of filters and settings can be used to explore which genes have been expressed in particular cells at specific points in time. This capability allows researchers to focus on specific cell populations or genes of interest.
"sCIRCLE is also a valuable communication tool for collaboratively examining data sets or presenting results," says Lars Barquist. Barquist, a computational biologist who initiated the study, leads a research group at the Helmholtz Institute Würzburg.
Barquist is also a professor at the University of Toronto in Canada. Additionally, sCIRCLE is already compatible with virtual reality devices, marking a step towards immersive 3D data visualization and interaction.
A future-oriented cross-disciplinary collaboration
The user-friendliness of sCIRCLE is notable. "The interface is easy to use and intuitively designed. sCIRCLE is therefore particularly helpful for biologists without extensive knowledge of bioinformatics," adds Barquist.
The designers and bioinformaticians worked together to make the data as simple and visually appealing as possible. "Collaboration between the disciplines of bioinformatics and design is extremely important as our data sets continue to grow and we need new ways to make them understandable," says Barquist.
To realize the project, THWS Master's student Maximilian Seeger spent some time in the HIRI research group. There, he had the opportunity to work directly with the scientists.
"This allowed me to understand what they wanted to explore in their data and develop an interface that would make this possible," said Maximilian Seeger. In test runs with data collected by HIRI researchers from different groups, the scientists tried out the tool and provided feedback.
"The joint development of sCIRCLE shows the importance of interdisciplinary and cross-institutional cooperation. I am very pleased we successfully combined the expertise of two Würzburg-based institutions in this project and look forward to future collaborations," says Erich Schöls, professor for interactive media at THWS, who supervised Max Seeger together with Lars Barquist.
"This is hopefully just the first step towards developing intuitive, interactive tools for data analysis," says Barquist. "We will build on this foundation to make our tools even more immersive and user-friendly."
While this application currently offers only basic virtual reality integration, the team plans to create additional easy-to-use virtual spaces for data exploration in the future.
More information: Maximilian Seege et al, s CIRCLE—An interactive visual exploration tool for single cell RNA-Seq data, NAR Genomics and Bioinformatics (2024). DOI: 10.1093/nargab/lqae084
Provided by Helmholtz Association of German Research Centres