This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
New tool developed to improve deadly superbug treatment
World-leading software that can both detect and predict antimicrobial resistance (AMR) in one of nature's toughest and deadliest superbugs has been developed by a University of the Sunshine Coast-led research team. The work is published in the journal Genome Medicine.
Pseudomonas aeruginosa is a common bacterium found in water and soil that can cause human infections ranging from swimmer's ear and skin inflammation to deadly pneumonia, sepsis, lung failure and bone disease.
A leading cause of bacterial mortality globally, the pathogen's innate resistance to many antibiotics, and its ability to develop resistance to newer antibiotics, makes it difficult to treat.
The software breakthrough follows six years of collaborative laboratory, clinical and bioinformatics work led by UniSC Center for Bioinnovation members Associate Professor Erin Price and Dr. Derek Sarovich and their UniSC Ph.D. graduate Dr. Danielle Madden at the Sunshine Coast Health Institute.
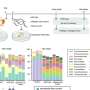
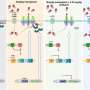
The innovative software uses whole-genome sequencing of the pathogen's DNA to identify the precise genetic changes that cause AMR.
This new ability to predict the pathogen's resistance to 10 of the most clinically important antibiotics will allow medical professionals to diagnose P. aeruginosa AMR infections earlier and more accurately, enabling faster and more effective treatment options.
Dr. Price, a molecular geneticist and microbiologist, said it was a potential game-changer in helping doctors decide what type of antibiotics to prescribe to patients with P. aeruginosa infection.
"Antimicrobial resistance is a global health epidemic that requires urgent mitigation to avoid a post-antibiotic era, where simple infections cannot be treated with antimicrobials that once revolutionized modern medicine," she said.
"If unmitigated, AMR infections are predicted to cause 10 million deaths globally by 2050 and cost more than $100 trillion a year.
"Earlier and more accurate diagnosis is crucial, particularly in such a dangerous and highly multi-drug-resistant pathogen.
"Our new software provides a rapid and highly accurate method for AMR identification in P. aeruginosa.
"It is already available to hospitals with onsite genome sequencing capacity, and hopefully this technology will also become routine in GP clinics in future.
"If our software becomes widely used, doctors could better target their antibiotic prescriptions, which in turn could lessen antibiotic overuse and slow AMR development across populations."
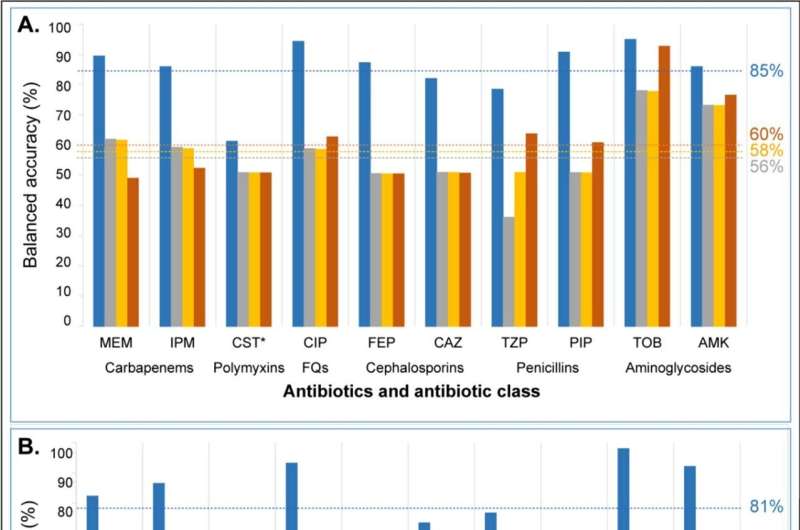
Dr. Sarovich, a molecular microbiologist and bioinformaticist, said the research found the new software tool was far superior to existing AMR detection software in terms of accuracy and precision.
"It produces results close to the current 'gold standard' in antimicrobial susceptibility testing, which is achieved through a slow, laborious method," he said.
"We tested the software in both Australian and international strain datasets, so it will have excellent use in diagnostic, public health, research, and pathogen surveillance laboratories both here and abroad.
"It could also be applied in agriculture, veterinary practice and environmental settings, as the pathogen also causes disease in plants and in horses, livestock and companion animals."
Dr. Madden said working on the ARDaP software project during her UniSC Ph.D. was inspiring.
"It was rewarding to see theoretical research applied directly to solve real-world problems," she said.
"The collaboration between laboratory researchers, clinicians and bioinformaticians was key to our success and highlights the importance of interdisciplinary approaches in tackling complex challenges like AMR."
More information: Danielle E. Madden et al, Keeping up with the pathogens: improved antimicrobial resistance detection and prediction from Pseudomonas aeruginosa genomes, Genome Medicine (2024). DOI: 10.1186/s13073-024-01346-z
Journal information: Genome Medicine
Provided by University of the Sunshine Coast