This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Using soil bacteria to help accelerate discovery of new antibiotics

Northeastern researcher Kim Lewis is spearheading an effort to accelerate discovery of new antibiotics as part of a multi-institutional project to tackle the growing problem of antibiotic resistance.
"It's a big issue because we largely stopped introducing novel antibiotics about 50 years ago," says Lewis, distinguished professor of biology and director of Northeastern's Antimicrobial Discovery Center.
"Bacteria continue to acquire and spread resistance, which has led to the antimicrobial resistance crisis the World Health Organization calls a slow-moving pandemic" that contributes to nearly 5 million deaths a year, Lewis says.
The goal of the accelerated technology is to use a high throughput approach and microfluidics to hasten the discovery of novel antibiotics.
Scientists from 25 research groups in nine states and the United Kingdom will develop novel tools, including single-cell assays and machine learning, to diagnose and treat bacterial infections proving resistant to current therapies.
"Existing antibiotics come from less than 1% of species out there," Lewis says. That means the vast majority of bacterial species have not been tested for their antimicrobial abilities.
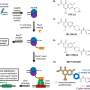
He and his team developed a new approach to discover antibiotics from soil bacteria that encapsulates single cells into microdroplets that can be tested at a rate of 10 million species a day instead of the historic rate of 100,000 species over the course of 10 years.
Lewis' research is part of a federal contract led by Johan Paulsson of Harvard Medical School. A microfluidics method developed by Paulsson places each cell in a channel of a microfluidic device, enabling visualization, which should further improve discovery of new antibiotics, Lewis says.
"So far, screening has been performed using early 20th-century technologies," he says. "Adopting this advanced screening platform will improve the chances to discover novel antibiotics."
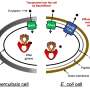
"The old-fashioned way is to take a petri dish and spread it evenly with a test pathogen. Then you put your potential producer on top of the petri dish. If it's making an antibiotic, you will see a zone of inhibition that separates the colony of your producer from the lawn of your test pathogen. That's been the standard technology."
"We're bringing this into the 21st century using microfluidics and rapid scanning laser microscopy that looks at 10 million cells more or less simultaneously and decides what's happening with them," Lewis says.
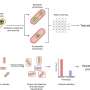
Relying on soil samples collected around the world but mainly in the U.S., "We make a suspension of cells from the environment. Then that suspension of cells enters into a microfluidic chip, which is sort of like a river, with side channels."
"There are approximately 10 million of such side channels. The suspension is diluted so that each channel gets approximately one cell that enters into the channel. Now that cell can start growing, making daughter cells and producing whatever it wants to produce."
Links connecting the tiny channels—which are only one one-thousandth of a millimeter—are opened to allow the antibiotic to diffuse to test pathogens.
Next, a scanning laser microscope "can tell us whether the test pathogen is dividing or not," Lewis says. "If the cell stopped growing, we know that it's being hit by an antibiotic."
Most antibiotics used in the clinic today were isolated from organisms called actinomycetes, which Lewis says were "over mined" long ago, leading to the antimicrobial resistance crisis.
He says his team at Northeastern has already started mining outside actinomycete bacteria and has found "very interesting new compounds that are now in development against multidrug resistance pathogens."
His lab has a track record of discovering new antibiotics, Lewis says, "and now we're going to do that better and faster."
Provided by Northeastern University
This story is republished courtesy of Northeastern Global News news.northeastern.edu.