This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Parasitic habit drives plastid genome structural variation and gene loss in Cuscuta species
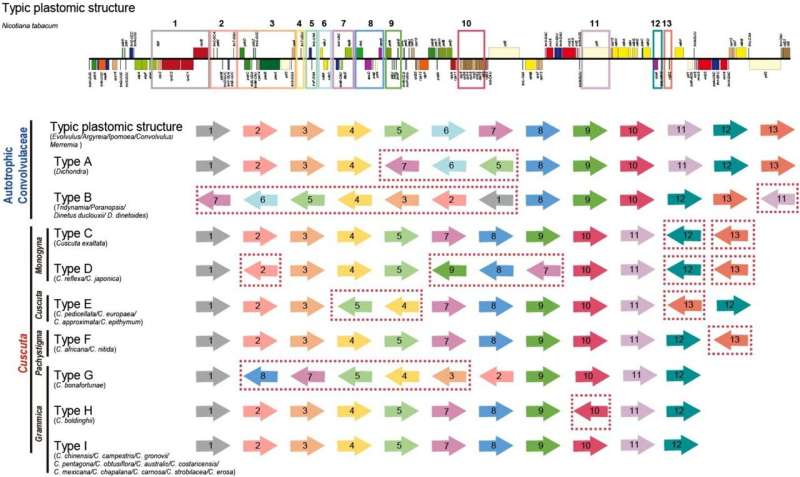
Hemiparasites obtain nutrients and inorganic salts from host plants through haustoria, a habit that has evolved independently at least 12 times in angiosperms. Cuscuta represents one of the 12 angiosperm orders that have independently evolved from autotrophs to parasites.
Morphologically, Cuscuta species are stem-hippophores, with roots and leaves completely degenerated or very weakly photosynthetically active or lost. Loss of plastid genes and genome functional degeneration have been reported several times in Cuscuta. However, the evolutionary links between its gene and genome functional degeneration and evolution have not yet been clarified.
Researchers from the Xishuangbanna Tropical Botanical Garden (XTBG) of the Chinese Academy of Sciences and their collaborators conducted de novo assembly of 29 new plastomes including 20 samples from seven Cuscuta species and nine autotrophic species of Convolvulaceae.
They deciphered the mechanism of plastome evolution in Cuscuta and its autotrophic plant relatives of Cuscutaceae. Their study is published in Plant Molecular Biology.
The results showed that that the structural variation of plastomes in Convolvulaceae was diverse, with nine types of structural rearrangements and six types of inverted repeat (IR) border expansion-contraction. The structural variations were closely related to the parasitic lifeform transition and may have exacerbated by IR border expansion-contraction and large repeat fragments.
In addition, plastome degeneration of Cuscuta species was progressive, with massive gene loss occurring only in three species from the Ceratophorae group of Grammica subgenus.
Overall, the parasitic habit of Cuscuta promoted the exposure of plastome genes to relaxed selective constraints, and the accumulation of microvariations in a large number of plastome genes led to plastome degeneration.
"Our study provides new evidence for a better understanding of plastome evolution, variation, and reduction in the genus Cuscuta," said Yu Wenbin of XTBG.
More information: Li-Qiong Chen et al, Variations and reduction of plastome are associated with the evolution of parasitism in Convolvulaceae, Plant Molecular Biology (2024). DOI: 10.1007/s11103-024-01440-1
Provided by Chinese Academy of Sciences

















