This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Study provides evidence for hidden diversity within Hydnoraceae
Researchers from the Wuhan Botanical Garden of the Chinese Academy of Sciences (CAS) have conducted a phylogenetic analysis of Hydnora plants in the family Hydnoraceae through plastid genome sequencing, assembly and annotation after extensive data querying and data collation. Their results are published in BMC Ecology and Evolution.
Hydnoraceae consists of two genera, Hydnora and Prosopanche. The former is distributed from southern Africa to the Arabian Peninsula and Madagascar, the latter from South to Central America. This holoparasitic family of root parasites is one of the oldest parasitic angiosperm lineages and belongs to the magnoliid order Piperales. Together with Aristolochiaceae, Asaraceae, and Lactoridaceae, they form the perianth-bearing clade within the order.
Hydnoraceae are unique among angiosperms due to their extreme loss of morphological features, such as the loss of leaves or scales, and their ability to emit a putrid odor that attracts beetles for pollination, hence their description as "the strangest plants on Earth."
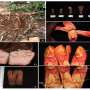
According to the researchers, three Hydnora species have been recorded in Kenya, namely H. sinandevu Beentje & Q. Luke, H. abyssinica A. Braun, and H. africana Thunb. These species are widely used for medicine, as a food source, and their roots are essential for tanning leather. Hydnora abyssinca has been described as having the widest distribution of any member of the family, ranging from southern Africa to northern East Africa.
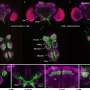
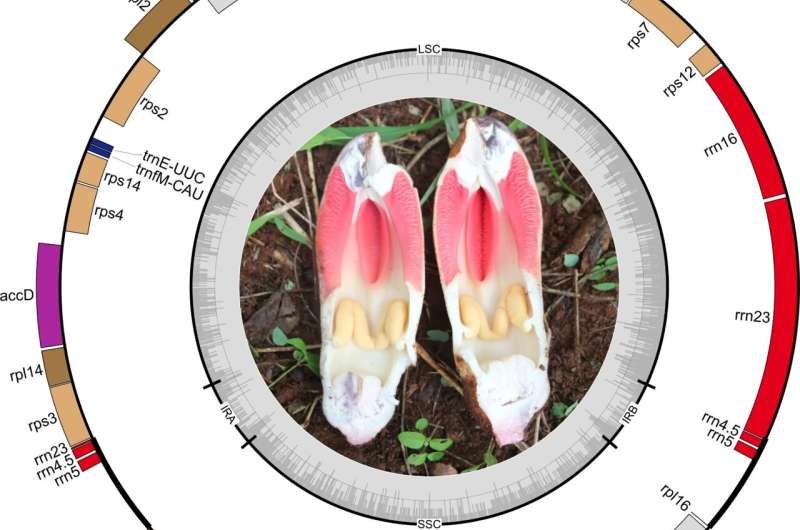
Here, for the first time, the researchers explored the interspecies plastome diversity in Hydnoraceae. They reported seven new plastomes of Hydnora abyssinica from Kenya and compared them with published plastomes of H. abyssinica from Namibia and H. arabica Bolin & Musselman from Oman. Based on the genome reconstruction and a resolved phylogenetic hypothesis including a broader sampling in Hydnoraceae, they analyzed their structure, nucleotide composition, and codon usage bias.
The researchers reported the first plastomes from Kenyan Hydnora abyssinica accessions. The plastomes have a typical quadripartite structure and encode 24 unique genes. Phylogenetic tree reconstruction recovered the Kenyan accessions as monophyletic and together in a clade with the Namibian H. abyssinica accession and the recently published H. arabica from Oman. Hydnora abyssinica as a whole, however, is recovered as non-monophyletic, with H. arabica nested within.
This result is supported by distinct structural plastome synapomorphies as well as pairwise distance estimates that reveal hidden diversity within the Hydnora species in Africa.
More information: Elijah Mbandi Mkala et al, Phylogenetic and comparative analyses of Hydnora abyssinica plastomes provide evidence for hidden diversity within Hydnoraceae, BMC Ecology and Evolution (2023). DOI: 10.1186/s12862-023-02142-w
Journal information: BMC Ecology and Evolution
Provided by Chinese Academy of Sciences