This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Research improves multiplex mutagenesis to increase experimental efficiency in plant genome editing

CRISPR/Cas9 remains the most powerful tool to generate mutations in plant genomes. Studying the various combinations of mutations has significantly increased the scale of experimental setups, necessitating more space to grow numerous plants.
Researchers from VIB-UGent Center for Plant Systems Biology have improved multiplex mutagenesis, which reduces the complexity and cost of large-scale genome editing projects. Their results have been published in The Plant Journal.
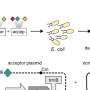
CRISPR/Cas experiments are continually increasing in scale, not only in terms of the number of mutants created through precise genome editing but also in terms of the number of genes that can be mutated simultaneously. The lab of Thomas Jacobs from VIB-UGent Center for Plant Systems Biology has developed screens to systematically mutate tens, hundreds, or even thousands of genes at a time.
The goal is to enhance the efficiency of inheritable germline mutations, and ultimately reduce the complexity and cost of large-scale genomic editing projects.
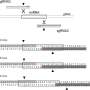
To achieve this, the team focused on two key aspects of CRISPR/Cas9 vector design: the promotor to drive Cas9 expression, and the nuclear localization signals (NLS) that direct the protein to the nucleus. By genotyping thousands of Arabidopsis plants, they found that using the RPS5A promotor to express Cas9 led to the highest mutation rate, and that flanking the Cas9 protein with bipartite NLS was the most efficient configuration to create germline mutations.
Combining these two elements results in the highest observed multiplex editing efficiency, with 99% of plants harboring at least one knockout mutation and over 80% with 4 to 7 mutations.
"This represents a significant advancement in the field of plant genetics and provides a reliable and efficient tool for researchers who focus on complex genetic engineering. What I find particularly interesting is the effect of the NLS. I daresay it had a stronger effect than the promote," said Dr. Thomas Jacobs, Group leader at VIB-UGent Center for Plant Systems Biology
The optimizations achieved in the study significantly reduce the complexity and cost of large-scale genome editing projects in plant science. To put it in numbers: with their previous vector, a CRISPR screen looking for all double knockouts of just 20 genes was estimated to require a population of about 18,000 plants. With the new vectors, it should take about 3,000 plants.
"These optimizations will be useful to generate higher-order knockouts in the germline of Arabidopsis and likely apply to other CRISPR systems as well," said Ward Develtere, Ph.D. student and lead author of the report.
More information: Ward Develtere et al, Continual improvement of CRISPR‐induced multiplex mutagenesis in Arabidopsis, The Plant Journal (2024). DOI: 10.1111/tpj.16785
Journal information: The Plant Journal
Provided by VIB (the Flanders Institute for Biotechnology)