This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
New time-resolved ultraviolet photodissociation mass spectrometry strategy for target protein stability analysis
How mutations impact protein stability and structure dynamics is crucial for understanding the molecular mechanism of the disease and the targeted drug design. However, probing the molecular details of mutation-induced subtle structure dynamics is still challenging.
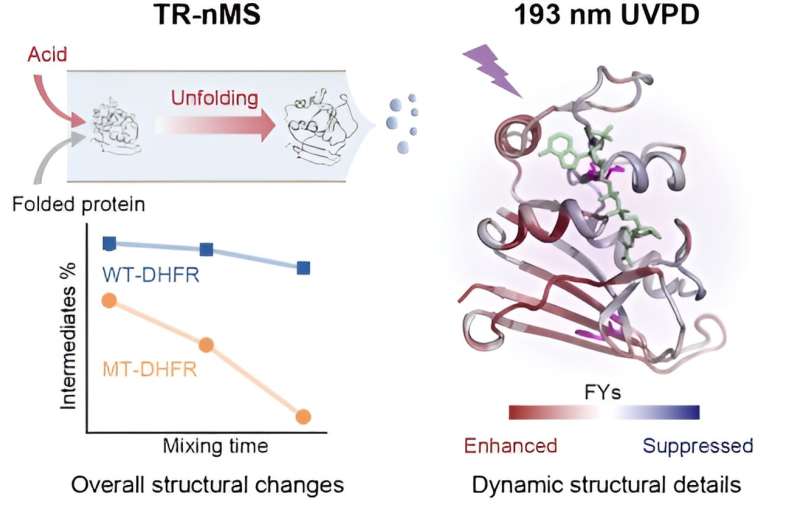
A research group led by Prof. Wang Fangjun from the Dalian Institute of Chemical Physics (DICP) of the Chinese Academy of Sciences has developed a time-resolved native mass spectrometry (TR-nMS) strategy coupled with ultraviolet photodissociation (UVPD) analysis. This strategy can interrogate the mutation-induced subtle alterations in protein stability and structure unfolding dynamics. The study is published in the Journal of the American Chemical Society.
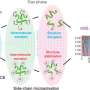
The researchers initiated the protein unfolding process by mixing the protein online with formic acid. With native mass spectrometry (nMS) and non-denaturing electrospray ionization (nESI), they monitored the species and relative intensities of acid-initiated protein unfolding intermediates via the unique charge state distributions (CSDs), and they quantitatively characterized the M42T/H114R mutations induced stability alterations of target proteins.
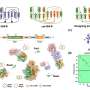
In addition, the researchers employed UVPD and fragment ion mass spectrometry methods to quantitatively compare the dynamic structure and molecular details of the unfolding intermediates of wild-type dihydrofolate reductase (DHFR) and the mutant.
The UVPD analysis revealed the special stabilization effect of cofactor nicotinamide adenine dinucleotide phosphate (NADPH) on DHFR structure, and that the M42T/H114R mutations could reduce the non-covalent NADPH-DHFR interactions, including residues I41, Q65, V78, D79, I82, and R98, thus promoting a decrease in stability.
"This work provides a new technique for studying the mutation-induced subtle structure dynamics and pathological mechanisms," said Prof. Wang.
More information: Pan Luo et al, Time-Resolved Ultraviolet Photodissociation Mass Spectrometry Probes the Mutation-Induced Alterations in Protein Stability and Unfolding Dynamics, Journal of the American Chemical Society (2024). DOI: 10.1021/jacs.4c00316
Journal information: Journal of the American Chemical Society
Provided by Chinese Academy of Sciences