This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Research team resolves decades-long problem in microscopy

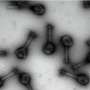
When viewing biological samples with a microscope, the light beam is disturbed if the lens of the objective is in a different medium than the sample. For example, when looking at a watery sample with a lens surrounded by air, the light rays bend more sharply in the air around the lens than in the water.
This disturbance leads to the measured depth in the sample being smaller than the actual depth. As a result, the sample appears flattened.
"This problem has been known for a long time, and since the 80s, theories have been developed to determine a corrective factor for determining the depth. However, all these theories assumed that this factor was constant, regardless of the depth of the sample. This happened despite the fact that the later Nobel laureate Stefan Hell pointed out in the 90s that this scaling could be depth-dependent," explains Associate Professor Jacob Hoogenboom from Delft University of Technology.
Calculations, experiments, and web tool
Sergey Loginov, a former postdoc at Delft University of Technology, has now shown with calculations and a mathematical model that the sample indeed appears more strongly flattened closer to the lens than farther away. Ph.D. candidate Daan Boltje and postdoc Ernest van der Wee subsequently confirmed in the lab that the corrective factor is depth-dependent.
The work is published in the journal Optica.
Last author Ernest Van der Wee says, "We have compiled our results into a web tool and software provided with the article. With these tools, anyone can determine the precise corrective factor for their experiment."
Understanding abnormalities and diseases
"Partly thanks to our calculation tool, we can now very precisely cut out a protein and its surroundings from a biological system to determine the structure with electron microscopy. This type of microscopy is very complex, time-consuming, and incredibly expensive. Ensuring that you are looking at the right structure is therefore very important," says researcher Daan Boltje.
"With our more precise depth determination, we need to spend much less time and money on samples that have missed the biological target. Ultimately, we can study more relevant proteins and biological structures. And determining the precise structure of a protein in a biological system is crucial for understanding and ultimately combating abnormalities and diseases."
In the provided web tool, you can fill in the relevant details of your experiment, such as the refractive indices, the aperture angle of the objective, and the wavelength of the light used. The tool then displays the curve for the depth-dependent scaling factor. You can also export this data for your own use. Additionally, you can plot the result in combination with the result of each of the existing theories.
More information: Sergey Loginov et al, Depth-dependent scaling of axial distances in light microscopy, Optica (2024). DOI: 10.1364/OPTICA.520595
Journal information: Optica
Provided by Delft University of Technology