This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Researchers reveal a hidden trait in Mycobacterium genomes governing stress adaptation
A new study, led by Qingyun Liu, Ph.D., assistant professor in the Department of Genetics, has uncovered a genetic feature known as "transcriptional plasticity," which plays a pivotal role in governing the transcriptional response of Mycobacteria to stressful conditions.
Bacterial cells must swiftly modulate the expression of their genes to cope with abrupt changes in the external environment. However, the extent to which certain genes can alter their expression in response to environmental shifts, as opposed to maintaining stable expression levels, has long puzzled scientists. Understanding how bacteria regulate these disparate transcriptional processes and the genetic features that underlie them has remained a challenge.
In a collaboration with researchers from UNC-Chapel Hill, Harvard, and Fudan University, lead researcher Qingyun Liu, Ph.D., set out to unravel the complex factors governing the transcriptional response in Mycobacterium tuberculosis (Mtb), the bacterial pathogen responsible for tuberculosis, which remains the leading cause of death due to a single infectious agent, with more than 10.6 million new cases and 1.6 million deaths each year.
Their study, titled "Genetically encoded transcriptional plasticity underlies stress adaptation in Mycobacterium tuberculosis," was published in the journal Nature Communications.
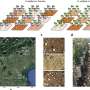
The researchers analyzed a comprehensive dataset comprising 894 RNA-Seq samples derived from 73 distinct conditions, which were generated in previous studies and curated by the researchers for meta-analysis purposes.
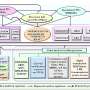
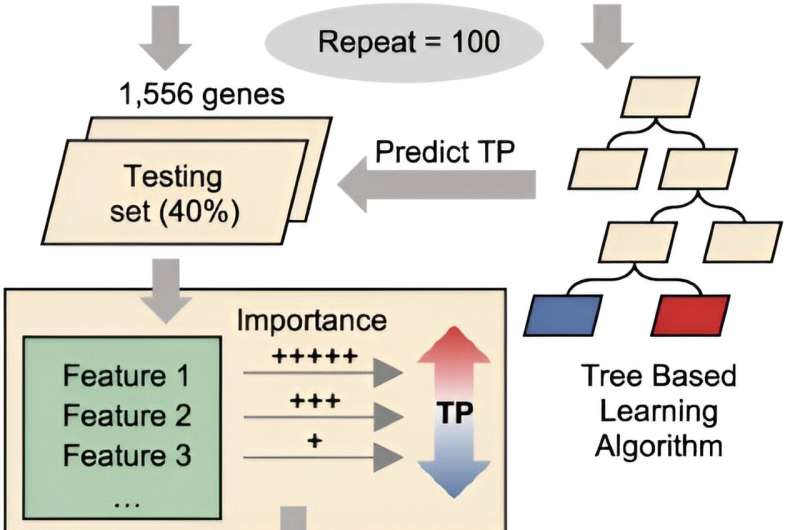
The researchers interrogated the transcriptional plasticity (TP) of each gene of Mtb, serving as a proxy for the variability of gene expression in response to environmental changes. Their analysis unveiled significant TP variation among Mtb genes, correlating with gene function and essentiality. Furthermore, they discovered that critical genetic features, such as gene length, GC content, and operon size, independently impose constraints on TP, extending beyond trans-regulation.
For instance, genes with shorter lengths generally exhibited higher TP compared to those with longer lengths. Additionally, genes with the lowest TP profiles were concentrated in a group with GC content aligning closely with the genome-wide average level (65%).
Liu said, "These features, previously not linked to transcriptional regulation in mycobacteria, are now recognized as factors that Mtb has evolved to shape its genes' TP."
Leveraging the genetic features identified as contributing to TP, the researchers were able to partially predict the TP levels of Mtb genes using a machine learning model. However, Liu pointed out that while this model shows promise, it is not yet perfect in predicting TP levels. This suggests that there may still be unidentified factors influencing TP that warrant further investigation.
By extending their analysis to include two other Mycobacteria species, namely M. smegmatis and M. abscessus, the researchers demonstrated a striking conservation of the TP landscape across different species of Mycobacteria, implying an evolutionary significance of TP as a conserved adaptive strategy among mycobacteria.
The researchers emphasized that TP can now serve as a useful supplement to gene essentiality and vulnerability for understanding bacterial physiological processes. This information can help prioritize gene candidates that can be targeted for drug purposes or mechanistic dissection.
Furthermore, the researchers showed that TP can function as a benchmark factor for future transcriptional studies, aiding in the identification of differentially expressed genes. This underscores the broader implications of TP in advancing our understanding of bacterial gene regulation and adaptation mechanisms.
More information: Cheng Bei et al, Genetically encoded transcriptional plasticity underlies stress adaptation in Mycobacterium tuberculosis, Nature Communications (2024). DOI: 10.1038/s41467-024-47410-5
Journal information: Nature Communications