This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Deciphering genomic language: New AI system unlocks biology's source code
Artificial intelligence (AI) systems like ChatGPT have taken the world by storm. There isn't much in which they're not involved, from recommending the next binge-worthy TV show to helping navigate through traffic. But can AI systems learn the language of life and help biologists reveal exciting breakthroughs in science?
In a new study published in Nature Communications, an interdisciplinary team of researchers led by Yunha Hwang, Ph.D. candidate in the Department of Organismic and Evolutionary Biology (OEB) at Harvard University, have pioneered an artificial intelligence (AI) system capable of deciphering the intricate language of genomics.
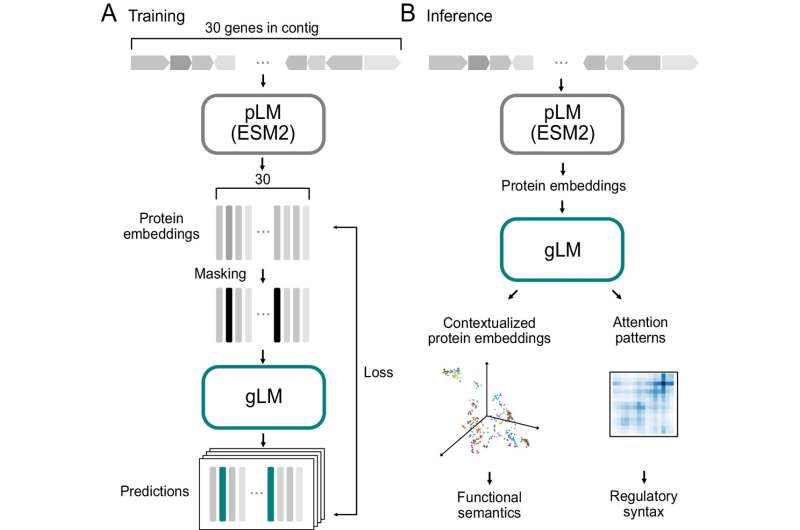
Genomic language is the source code of biology. It describes the biological functions and regulatory grammar encoded in genomes. The researchers asked, "Can we develop an AI engine to 'read' the genomic language and become fluent in the language, understanding the meaning, or functions and regulations, of genes?" The team fed the microbial metagenomic data set, the largest and most diverse genomic dataset available, to the machine to create the Genomic Language Model (gLM).
"In biology, we have a dictionary of known words and researchers work within those known words. The problem is that this fraction of known words constitutes less than one percent of biological sequences," said Hwang. "The quantity and diversity of genomic data is exploding, but humans are incapable of processing such a large amount of complex data."
Large language models (LLMs), like GPT4, learn meanings of words by processing massive amounts of diverse text data that enables understanding the relationships between words. The Genomic Language Model (gLM) learns from highly diverse metagenomic data, sourced from microbes inhabiting various environments including the ocean, soil and human gut.
With this data, gLM learns to understand the functional "semantics" and regulatory "syntax" of each gene by learning the relationship between the gene and its genomic context. gLM, like LLMs, is a self-supervised model—this means that it learns meaningful representations of genes from data alone and does not require human-assigned labels.
Researchers have sequenced some of the most commonly studied organisms like people, E. coli, and fruit flies. However, even for the most studied genomes, the majority of the genes remain poorly characterized.
"We've learned so much in this revolutionary age of 'omics,' including how much we don't know," said senior author Professor Peter Girguis, also in OEB at Harvard. "We asked, how can we glean meaning from something without relying on a proverbial dictionary? How do we better understand the content and context of a genome?"
The study demonstrates that gLM learns enzymatic functions and co-regulated gene modules (called operons), and provides genomic context that can predict gene function. The model also learns taxonomic information and context-dependencies of gene functions.
Strikingly, gLM does not know which enzyme it is seeing, nor which bacteria from which the sequence comes. However, because it has seen many sequences and understands the evolutionary relationships between the sequences during training, it is able to derive the functional and evolutionary relationships between sequences.
"Like words, genes can have different 'meanings' depending on the context they are found in. Conversely, highly differentiated genes can be 'synonymous' in function. gLM allows for a much more nuanced framework for understanding gene function. This is in contrast to the existing method of one-to-one mapping from sequence to annotation, which is not representative of the dynamic and context-dependent nature of the genomic language," said Hwang.
Hwang teamed with co-authors Andre Cornman (an independent researcher in machine learning and biology), Sergey Ovchinnikov (former John Harvard Distinguished Fellow and current Assistant Professor at MIT), and Elizabeth Kellogg (Associate Faculty at St. Jude Children's Research Hospital) to form an interdisciplinary team with strong backgrounds in microbiology, genomes, bioinformatics, protein science, and machine learning.
"In the lab we are stuck in a step-by-step process of finding a gene, making a protein, purifying it, characterizing it, etc. and so we kind of discover only what we already know," Girguis said. gLM, however, allows biologists to look at the context of an unknown gene and its role when it's often found in similar groups of genes. The model can tell researchers that these groups of genes work together to achieve something, and it can provide the answers that do not appear in the "dictionary."
"Genomic context contains critical information for understanding the evolutionary history and evolutionary trajectories of different proteins and genes," Hwang said. "Ultimately, gLM learns this contextual information to help researchers understand the functions of genes that previously were unannotated."
"Traditional functional annotation methods typically focus on one protein at a time, ignoring the interactions across proteins. gLM represents a major advancement by integrating the concept of gene neighborhoods with language models, thereby providing a more comprehensive view of protein interactions," stated Martin Steinegger (Assistant Professor, Seoul National University), an expert in bioinformatics and machine learning, who was not involved in the study.
With genomic language modeling, biologists can discover new genomic patterns and uncover novel biology. gLM is a significant milestone in interdisciplinary collaboration driving advancements in the life sciences.
"With gLM we can gain new insights into poorly annotated genomes," said Hwang. "gLM can also guide experimental validation of functions and enable discoveries of novel functions and biological mechanisms. We hope gLM can accelerate the discovery of novel biotechnological solutions for climate change and bioeconomy."
More information: Yunha Hwang et al, Genomic language model predicts protein co-regulation and function, Nature Communications (2024). DOI: 10.1038/s41467-024-46947-9
Journal information: Nature Communications
Provided by Harvard University