This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
proofread
Innovative computational tools provide new insights into the polyploid wheat genome
A new review led by Associate Professor Weilong Guo (College of Agronomy and Biotechnology, China Agricultural University, Beijing, China) explores the polyploid wheat genome through new computational tools. The research is published in the journal aBIOTECH.
Bread wheat is allohexaploid, and its genome is about 16 Gb in size, one of the largest genomes in crops. Analyzing such massive genomic data, such as identifying genetic variations and understanding gene functions, requires high costs as well as robust and efficient computational tools.
In addition, polyploidization, frequent genomic variants, and intricate genomic resources further limit the study of wheat functional genomics. These challenges call for the design and development of novel computational tools for tackling complex genomes.
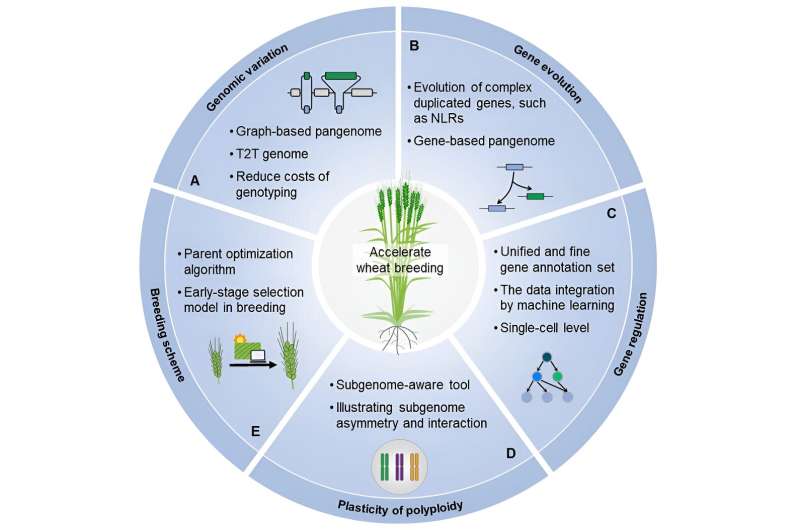
By targeting the unique features and challenges of the polyploid wheat genome and dissecting data from multiple omics levels, the researchers have recently developed a series of genomic tools to unveil genomic variation and gene function. The genomic databases or web servers were further developed for users to conveniently access these data without programming.
Additionally, a series of step-by-step examples are provided to illustrate how these tools or databases can be utilized for dissecting wheat germplasm resources and unveiling functional genes and regulations associated with important agronomic traits. The pipelines and commands for the step-by-step examples are available at http://wheat.cau.edu.cn/resources/tutorial.html.
The review also discusses the future potential innovations in designing genomic algorithms, developing tools, and constructing a database for crops with complex genomes, in terms of genomic variation, gene evolution, gene regulation, plasticity of polyploid, and breeding by design. The utilization of efficient and high-performance tools and databases will drive a transformation in wheat breeding technologies, thereby enhancing the development of superior wheat varieties to meet the increasing food demand of the growing population under ever-changing climate conditions.
More information: Yongming Chen et al, Innovative computational tools provide new insights into the polyploid wheat genome, aBIOTECH (2024). DOI: 10.1007/s42994-023-00131-7
Provided by Beijing Zhongke Journal Publising Co. Ltd.




















