This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Self-teaching web app improves speed, accuracy of classifying DNA variations among cereal varieties
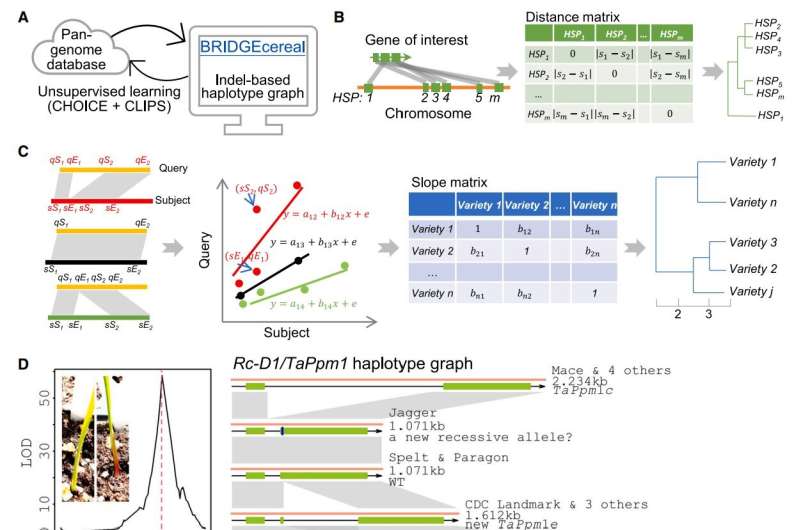
Agricultural Research Service and Washington State University scientists have developed an innovative web app called BRIDGEcereal that can quickly and accurately analyze the vast amount of genomic data now available for cereal crops and organize the material into intuitive charts that identify patterns locating genes of interest.
With the rapid advancements in the field of genomics the past 25 years, a game-changer for crop improvement has emerged referred to as the pan-genome, defined as the assembled genome sequences from multiple varieties within a species. But understanding and enhancing crops based on the huge amount of data that have been generated also has created a challenge for researchers due to the lack of efficient and user-friendly bioinformatic tools, particularly ones designed to handle large volume DNA variations in a species.
Take wheat, for example. The standard reference wheat genome—which was done for the wheat variety Chinese Spring—is five times larger than the human genome. In addition, researchers have long struggled with the wide variation in the locations of genes that control essential agronomic traits across wheat's 21 chromosomes. Right now, a dozen wheat genomes are publicly available.
This adds up to a huge amount of data, making analysis of it a tedious process even for researchers with advanced bioinformatic skills. It is particularly challenging to sort through all of the data to identify similar stretches of DNA that may control the same trait no matter where they are located on a chromosome.
BRIDGEcereal is designed to transform the process of identifying large DNA variation from tedious to efficient.
"By simply providing BRIDGEcereal with the sequence of DNA you are interested in, it will complete the search process in less than one minute," explained ARS research biologist Xianran Li, leader of the BRIDGEcereal project. Li is with the ARS Wheat Health, Genetics, and Quality Research Unit in Pullman, Washington.
"And BRIDGEcereal will organize the data it finds and present it to you in easily understood charts that highlight any patterns of where that DNA is," Li added.
It only took a minute for BRIDGEcereal to identify a promising candidate gene as the controller of a wheat mutation that reduces the length of awns, the bristle-like extensions from the wheat grain head. It had been known since the 1940s that a gene on wheat chromosome 4A controls awn development, which is an iconic wheat trait. But the exact gene controlling that trait has remained unknown.
"By searching dozens of potential genes through BRIDGEcereal, we were able to quickly identify a gene with a large DNA variation as the one that has been eluding researchers," Li said.
The scientists also designed BRIDGEcereal to be self-teaching—also called unsupervised machine-learning—meaning BRIDGEcereal can autonomously learn to recognize new patterns without the need for explicit instructions to follow.
"So what we've developed is a one-stop gateway to efficiently mine publicly accessible cereal pan-genomes that will only get more efficient as the data continues to mount up," Li said.
Bosen Zhang, a postdoctoral research associate with Washington State University and co-developer of the web app, added, "Researchers will find BRIDGEcereal to be an invaluable tool for selecting and prioritizing candidate genes that control specific traits in cereal crops."
BRIDGEcereal was first developed to work with wheat. It has already been adapted to analyze similar data from barley, maize, sorghum, and rice.
This research was published in the journal Molecular Plant.
More information: Bosen Zhang et al, Streamline unsupervised machine learning to survey and graph indel-based haplotypes from pan-genomes, Molecular Plant (2023). DOI: 10.1016/j.molp.2023.05.005
Journal information: Molecular Plant
Provided by United States Department of Agriculture