This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
DNA nanotube rings: Research team develops important building block for artificial cells
During cell division, a ring forms around the cell equator, which contracts to divide the cell into two daughter cells. Together with researchers from Heidelberg, Dresden, Tübingen and Harvard, Professor Jan Kierfeld and Lukas Weise from the Department of Physics at TU Dortmund University have succeeded for the first time in synthesizing such a contractile ring with the help of DNA nanotechnology and uncovering its contraction mechanism. The results have been published in Nature Communications.
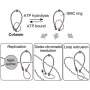
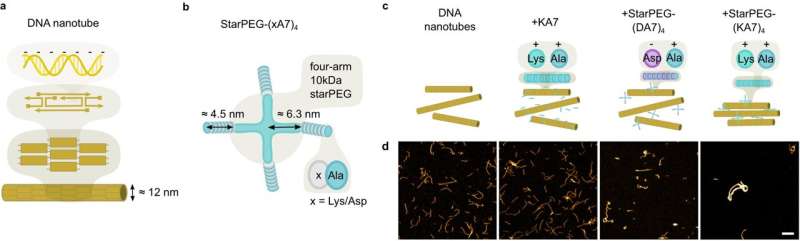
In synthetic biology, researchers try to recreate crucial mechanisms of life in vitro, such as cell division. The aim is to be able to synthesize minimal cells. The research team led by Professor Kerstin Göpfrich from Heidelberg University has now synthetically reproduced contractile rings for cell division using polymer rings composed of DNA nanotubes.
The formation of a ring that constricts and separates dividing cells is an important step in natural cell division. In nature, this is achieved by a machinery of proteins: motor proteins powered by chemical energy from ATP hydrolysis pull together a ring of filaments of the protein actin. Adenosine triphosphate, or ATP, is a molecule that occurs in all living cells and supplies the energy for numerous cellular processes.
The contraction mechanism of the DNA rings developed by the researchers no longer relies on motor proteins powered by ATP hydrolysis. Instead, molecular attraction between ring segments can trigger the contraction of the polymer rings.
This molecular attraction can be induced in two ways: either by crosslinking molecules with two "sticky" ends that can connect two polymer segments, or by means of the depletion interaction, where the polymers are surrounded by "crowder" molecules that press the segments together. This mechanism consumes no chemical energy, meaning that no energy source needs to be incorporated in the synthetic cell for the mechanism to function.
Professor Jan Kierfeld, Professor of Theoretical Physics, and doctoral researcher Lukas Weise are working in the field of biological physics. As part of their research work, they have developed a theoretical description and a molecular dynamics simulation of the contraction mechanism, which match the experimental results of their research partners.
To this end, they devised special methods for simulating the DNA rings on a realistic scale. Theory and the simulation make it possible to explain quantitatively how the polymer rings form and contract.
"This means we are able not only to predict that an increased concentration of 'crowder' molecules will make the ring smaller but also by how much smaller," says Professor Kierfeld. In this way, it is possible to determine how the diameter of the DNA ring can be precisely controlled, which is highly significant for future applications of contractile rings in synthetic biology.
Mechanisms for cell division are an important step towards an artificial cell, the construction of which facilitates a better understanding of the functional mechanisms of natural cells and, thus, of the foundations of life.
More information: Maja Illig et al, Triggered contraction of self-assembled micron-scale DNA nanotube rings, Nature Communications (2024). DOI: 10.1038/s41467-024-46339-z
Journal information: Nature Communications
Provided by TU Dortmund University