This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
New technique detects distinct fish populations in a single lake through their environmental DNA
Using the tiny fragments of DNA that animals leave behind as they move through an environment is giving researchers an ever-greater insight into where and how they live.
A new study has allowed scientists to not only see what species of fish are present in a lake, but also identify how populations of those fish differ.
The development of testing water for environmental DNA (eDNA) has allowed scientists and conservationists to know what species has recently been living in or moving through a specific area.
But the technique has its limitations. Typically, researchers test for a type of DNA known as mitochondrial DNA. While this is useful for giving a binary yes or no to the presence or absence of certain species within a sample of, say, lake water, it is often not precise enough to give more detailed information about the populations of those species.
To do this, researchers need to look for what is known as nuclear DNA. The issue here is that each nuclear genome is much larger but less abundant than mitochondrial DNA. This makes any given fragment of nuclear eDNA much rarer in the environment and more difficult to isolate.
But if it can be done, the potential could be huge. For example, it would allow researchers to track complex genetic changes happening within and between populations of animals without ever having to see the actual species.
This is where Dr. Rupert Collins, Senior Curator of Fishes at the Natural History Museum comes in, with his colleagues at the University of Bristol, University of Cambridge and the Tanzania Fisheries Research Institute. They have managed to detect the nuclear eDNA of two different populations of fish living within the same lake.
"I'm still amazed that it worked so well," says Rupert. "It was very much a pie in the sky project."
The results have been published in iScience.
Nature's laboratory
The myriad of lakes that are scattered across the rift valley of Eastern Africa are often referred to as a natural laboratory.
This is because, in a similar way to how the Galapagos islands isolated populations of birds, which led to the creation of new species, the lakes act like islands of isolation for fishes. This is particularly true for a group of fish known as the cichlids.
The continual geological upheaval of the region causing the crust to split apart and form countless lakes and rivers over this time has separated fish populations, brought others together, while causing the demise of yet more. This dynamic process has resulted in these lakes supporting at least 1,500 different species of cichlids, although even this is most likely a substantial underestimate.
But it is not just between lakes that there are differences in fish populations. Even within lakes, there can be multiple habitats that allow for fishes to gradually become different from each other and evolve into multiple species.
"We get this similar pattern in freshwater fishes of the northern hemisphere too," explains Rupert. "When all the ice sheets retreated and lakes formed, evolution happened quickly as new habitats were exposed."
"So it's not something specific to the tropics. It's happening right on our doorstep, but with chars, sticklebacks and whitefishes instead of cichlids."
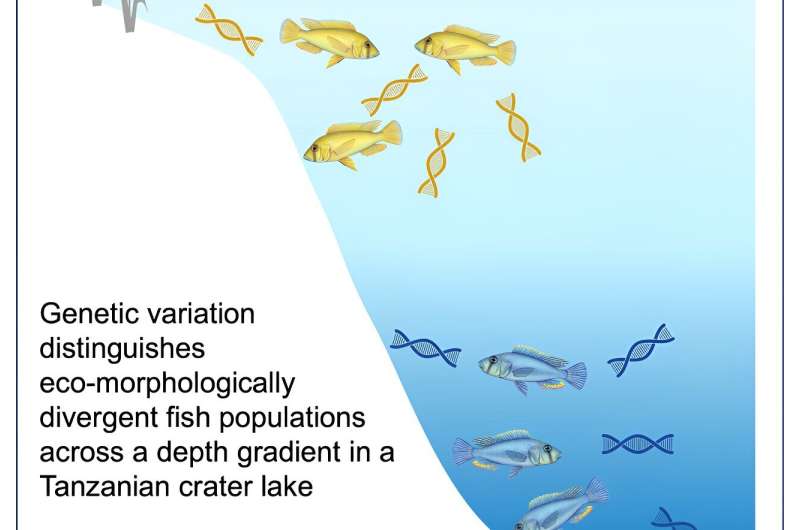
Lake Masoko in Tanzania is a great example of how this can occur. An almost perfectly circular crater lake, it plunges to around 35 meters in depth. Within it is a species of cichlid called Astatotilapia calliptera, which has divided into two populations within the last 1,000 years. For the fish that live in the shallower margins of the lake, the males are typically yellow in color and feed on benthic invertebrates, while those that live in the deeper water are instead a deep blue and feed on floating zooplankton.
Despite living in different places of the lake, looking and behaving differently, the two populations are perhaps not quite genetically distinct enough to be deemed separate species (although they may well be on that path).
But this means that the fish were the perfect subject for a study looking to see if it was possible to detect genetic variation within a species by sampling the water alone.
Testing the waters
Rupert and his colleagues took samples of water from different depths of the lake, ranging from the surface to close to the bottom. The water was then filtered to isolate any DNA it contained.
The researchers then analyzed the eDNA extracted from the water at different depths, and tested it to look for the specific genetic variation that is more likely to be found in either the shallower yellow fish or the deeper blue fish.
Over a distance of just 20 meters, the team found that the genetic variation associated with the yellow fish was more prevalent in the shallower waters and the genetic variation linked to the blue fish was more commonly detected in the deeper water.
"We compared the eDNA to sequenced genomes of each population, and we found a positive association at each depth," says Rupert. "The depth pattern is there, which is amazing."
This shows for the first time how the variation seen within a species can be detected by sampling nuclear eDNA over incredibly short distances. While Lake Masoko is quite a contained environment, the work is at least a proof of concept that sampling for nuclear eDNA is possible and that it could have further applications.
One such situation could be with cod, explains Rupert. As commercial fishing has intensified, and climate change has warmed the waters of Europe, southern populations of this cold-adapted species have been doing poorly, while the more northerly populations are doing better. But there is some evidence that populations are adjusting, and this is when eDNA could come in.
"We could, for example, target specific genes that are involved in adapting to warmer waters," explains Rupert. "We could see what entire cod populations are doing by looking at the water samples without actually catching any fish."
"That is the theory. But there is a big difference between doing it in a little lake and doing it in the sea."
With any hope, further refinement and development of the techniques should allow for researchers and conservationists to get an ever-clearer picture of how difficult-to-study animals are faring.
More information: Zifang Liu et al, Nuclear environmental DNA resolves fine-scale population genetic structure in an aquatic habitat, iScience (2023). DOI: 10.1016/j.isci.2023.108669
Provided by Natural History Museum
This story is republished courtesy of Natural History Museum. Read the original story here.