This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Researchers develop protocol for rapid detection of antibiotic-resistant bacteria

A paper-based platform developed by researchers at the Indian Institute of Science (IISc) and Jawaharlal Nehru Center for Advanced Scientific Research (JNCASR) could help quickly detect the presence of antibiotic-resistant, disease-causing bacteria.
One of mankind's greatest challenges has been the rise of disease-causing bacteria that are resistant to antibiotics. Their emergence has been fueled by the misuse and overuse of antibiotics.
A handful of such bacteria—including E. coli and Staphylococcus aureus—have caused over a million deaths, and these numbers are projected to rise in the coming years, according to the World Health Organization. Timely diagnosis can improve the efficiency of treatment.
"Generally, the doctor diagnoses the patient and gives them medicines. The patient then takes it for 2–3 days before realizing that the medicine is not working and goes back to the doctor. Even diagnosing that the bacteria is antibiotic-resistant from blood or urine tests takes time. We wanted to reduce that time-to-diagnosis," says Uday Maitra, Professor at the Department of Organic Chemistry, IISc.
In a paper published in ACS Sensors, Maitra's lab and collaborators have addressed this challenge. They have developed a rapid diagnosis protocol that uses a luminescent paper-based platform to detect the presence of antibiotic-resistant bacteria.
There are different ways by which a bacterium becomes resistant to antibiotics. In one, the bacterium evolves, and can recognize and eject the medicine out of its cell. In another, the bacterium produces an enzyme called β-lactamase, which hydrolyses the β-lactam ring—a key structural component of common antibiotics like penicillin and carbapenem—rendering the medication ineffective.
The approach developed by the IISc and JNCASR team involves incorporating biphenyl-4-carboxylic acid (BCA) within a supramolecular hydrogel matrix containing terbium cholate (TbCh). This hydrogel normally emits green fluorescence when UV light is shined on it.
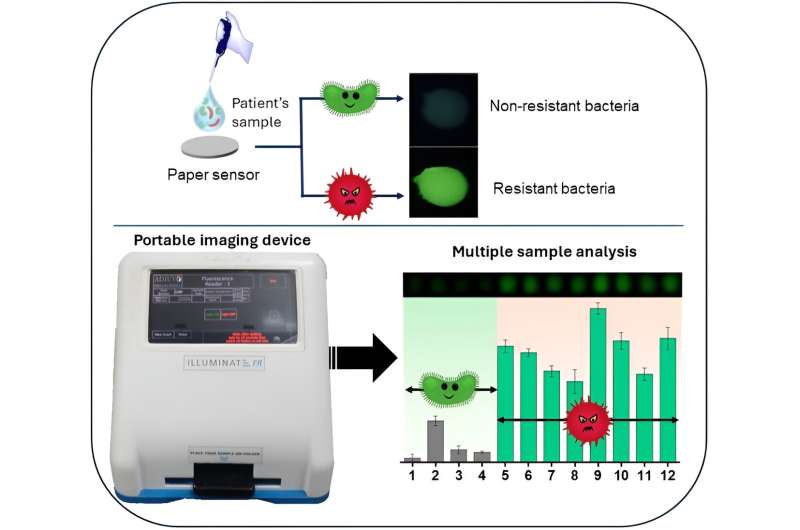
"In the lab, we synthesized an enzyme-substrate by tethering BCA to the cyclic [β-lactam] ring that is a part of the antibiotic. When you mix this with TbCh hydrogel, there is no green emission as the sensitizer is 'masked,'" explains Arnab Dutta, Ph.D. student in the Department of Organic Chemistry, IISc, and lead author of the paper.
"In the presence of β-lactamase enzyme, the gel will produce green emission. β-lactamase enzyme in the bacteria is the one that cuts open the drug, destroys, and unmasks the sensitizer BCA. So, the presence of β-lactamase is signaled by green emission."
The luminescence signals the presence of antibiotic-resistant bacteria, and the intensity of the luminescence indicates the bacterial load. For non-resistant bacteria, the green intensity was found to be extremely low, making it easier to distinguish them from resistant bacteria.
The next step was to find a way to make the technology inexpensive. Currently used diagnostics instruments are costly, which drives up the price for testing.
The team collaborated with a Tamil Nadu-based company called Adiuvo Diagnostics to design a customized, portable and miniature imaging device, named Illuminate Fluorescence Reader. Infusing the hydrogel in a sheet of paper as the medium reduced the cost significantly. The instrument is fitted with different LEDs that shine UV radiation as required. Green fluorescence from the enzyme is captured by a built-in camera, and a dedicated software app measures the intensity, which can help quantify the bacterial load.
The team from IISc tied up with Jayanta Haldar's research group from JNCASR to check their approach on urine samples. "We used samples from healthy volunteers and added pathogenic bacteria to mimic Urinary Tract Infections. It successfully produced the outcome within two hours," explains Maitra.
As the next step, the researchers plan to tie up with hospitals to test this technology with samples from patients.
More information: Arnab Dutta et al, Augmenting Antimicrobial Resistance Surveillance: Rapid Detection of β-Lactamase-Expressing Drug-Resistant Bacteria through Sensitized Luminescence on a Paper-Supported Hydrogel, ACS Sensors (2023). DOI: 10.1021/acssensors.3c02065
Journal information: ACS Sensors
Provided by Indian Institute of Science