This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
proofread
New technique for studying membrane-associated intrinsically disordered proteins
Intrinsically disordered proteins (IDPs) are widely found in the proteomes of eukaryotes and play key roles in life processes such as transcription of genetic information and signaling. Apart from being usually highly repetitive, hydrophilic and electrically charged as well as encoding simple sequences of genes, IDPs also distinguish themselves in their natural abundance and structural aspects, which become the basis of the "disorder–function paradigm" of proteins.
Over the last two decades, the role of IDPs in human diseases and as drug targets has been actively studied, while how to characterize the highly flexible and heterogeneous conformations of IDPs at high resolution remains a key issue in this field.
More than 15% of IDP molecules are membrane-bound in cells, and their internal dynamics and overall (translational and rotational) motions within the phospholipid bilayers are closely related to their physicochemical properties and biological functions, but these dynamic processes are difficult to capture and quantitatively characterized by conventional structural analysis methods.
A group of researchers led by Prof. Long Dong from the University of Science and Technology of China (USTC) has developed an IDP spectroscopy method based on the membrane paramagnetic relaxation enhancement (mPRE) technique, which has successfully achieved high-precision modeling of the internal conformations, orientations, and immersion depths of IDPs. The results have been published in the Journal of the American Chemical Society.
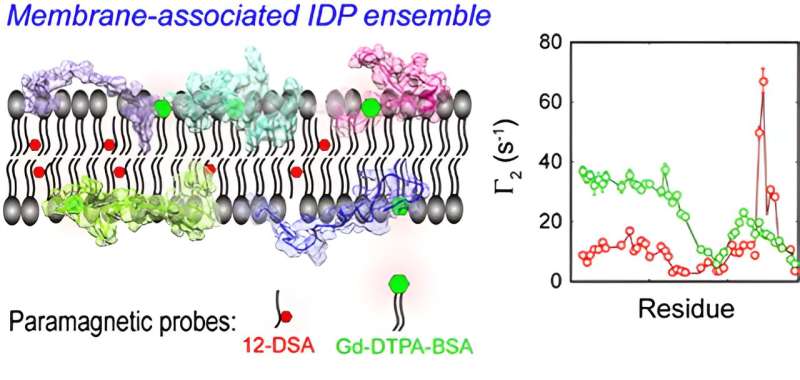
In this work, the researchers explored in detail the flexibility and mobility of the spin probe molecules within the membrane for accurate interpretation of the mPRE spectral data, and proposed a weighted three-dimensional (3D) grid model based on all-atom simulations for quantitatively portraying the effect of the spin probe dynamics on the membrane paramagnetic relaxation enhancement rate.
Taking advantage of the high computational efficiency of the model, the researchers further developed an algorithm which is detailed in the Supporting Information (SI) for optimizing orientations of the global and internal degrees of freedom of motion of membrane-bound IDPs via superposition of z-coordinates only, tailored for the mPRE data analysis, constructing an all-atom ensemble model of IDP in an implicit membrane environment.
CD3ε is a component of the T-cell receptor (TCR) complex responsible for T-cell antigen recognition. The CD3ε cytoplasmic domain (CD3εCD) contains immunoreceptor tyrosine-based activation motifs (ITAMs), and it forms a fuzzy complex with lipid bilayers in an intrinsically disordered state, and regulates the signaling activity of fuzzy complex by utilizing dynamic membrane shielding of key tyrosine sites.
The researchers resolved the ensemble based on molecular dynamics of CD3εCD in lipid bilayers by applying an all-atom ensemble model solution of IDP in an implicit membrane environment.
The ensemble from mPRE experimental parameters maps the dynamic distribution of CD3εCD in different regions of the membrane at the atomic level and reveals key differences in the membrane interactions of different tyrosine sites on ITAM, providing a new mechanistic explanation for the monophosphorylation pattern of ITAM.
The mPRE spectroscopic analysis method established in this work is expected to broadly facilitate atomic resolution studies of various functional membrane IDPs.
More information: Hong Jin et al, Quantitative Ensemble Interpretation of Membrane Paramagnetic Relaxation Enhancement (mPRE) for Studying Membrane-Associated Intrinsically Disordered Proteins, Journal of the American Chemical Society (2023). DOI: 10.1021/jacs.3c10847
Journal information: Journal of the American Chemical Society
Provided by University of Science and Technology of China