This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Scientists discover why chicken farms are a breeding ground for antibiotic resistant bacteria
Scientists from the University of Nottingham are one step closer to understanding how bacteria, such as E. coli and Salmonella enterica, share genetic material which makes them resistant to antibiotics.
Antimicrobial resistance (AMR), the capability of organisms to be resistant to treatment with antibiotics and other antimicrobials, is now one of the most threatening issues worldwide. Livestock farms, their surrounding environments and food products generated from husbandry, have been highlighted as potential sources of resistant infections for animals and humans.
In livestock farming, the misuse and overuse of broad-spectrum antimicrobials administered to reduce production losses is a major known contribution to the large increase and spread of AMR.
In this latest study, scientists provide a significant contribution to demonstrating that different bacteria species, co-existing within the same microbial community (for example, within the chicken gut), are able to share AMR-associated genetic material and end-up implementing similar resistance mechanisms. The discovery has important implications as it affects our understanding of AMR and poses further challenges to the implementation of solutions for surveillance and treatment/control.
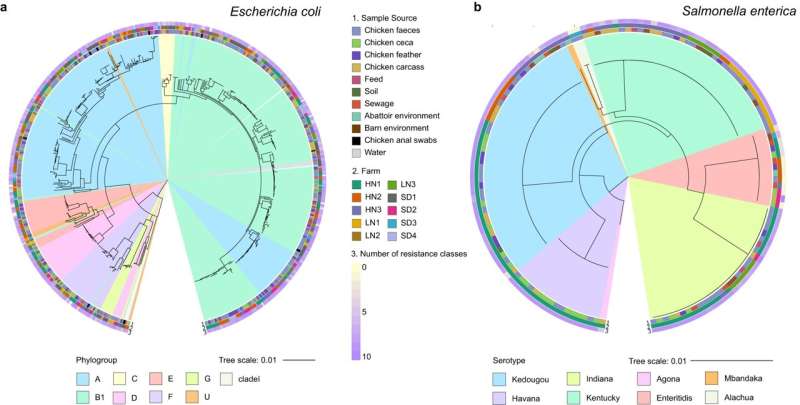
This study, published in Nature Communications, looks at two important bacteria found in food animals—Escherichia coli and Salmonella enterica, which both show high levels of drug resistance, are common in farming settings, have high levels of transmissibility to humans and cause food poisoning.
The research is a collaboration between experts from the University's School of Veterinary Medicine and Science, the China National Center for Food Safety Risk Assessment, New Hope Liuhe Group Ltd in China and Nimrod Veterinary Products Limited.
Dr. Tania Dottorini, from the School of Veterinary Medicine and Science at the University of Nottingham, is the lead researcher on the study. She said, "These species of bacteria can share genetic material both within, and potentially between species, a way in which AMR is spread. That is why understanding the extent to which these bacteria within the same environment, and importantly, the same host, can co-evolve and share their genome could help the development and more efficient treatments to fight AMR."
The team collected 661 E. coli and Salmonella bacteria isolates from chickens and their environments in 10 Chinese chicken farms and four abattoirs over a two-and-a-half-year period. They carried out a large-scale analysis using conventional microbiology DNA sequencing and data-mining methods powered by machine learning.
This is the first study of its kind where the genomic content of two bacteria species is characterized over such a large scale, using samples collected from the same animals, at the same time and from real-world settings (farms and abattoirs). The main findings indicate that E. coli and Salmonella enterica co-existing in the chicken gut, compared to those existing in isolation, feature a higher share of AMR-related genetic material, implement more similar resistance and metabolic mechanisms, and are likely the result of a stronger co-evolution pathway.
Dr. Dottorini says, "The insurgence and spread of AMR in livestock farming is a complex phenomenon arising from an entangled network of interactions happening at multiple spatial and temporal scales and involving interchanges between bacteria, animals and humans over a multitude of connected microbial environments.
"Investing in data mining and machine learning technologies capable to cope with large scale, heterogeneous data is crucial to investigate AMR , in particular when considering the interplay between cohabiting bacteria, especially in ecological settings where community-driven resistance selection occurs.
"Overall, this work has also demonstrated that the investigation of individual bacterial species taken in isolation may not provide a sufficiently comprehensive picture or the mechanisms underlying insurgence and spread of AMR in livestock farming, potentially leading to an underestimation of the threat to human health."
More information: Michelle Baker et al, Convergence of resistance and evolutionary responses in Escherichia coli and Salmonella enterica co-inhabiting chicken farms in China, Nature Communications (2024). DOI: 10.1038/s41467-023-44272-1
Journal information: Nature Communications
Provided by University of Nottingham