This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Molecular dynamics database offers improved understanding of COVID-19 proteins
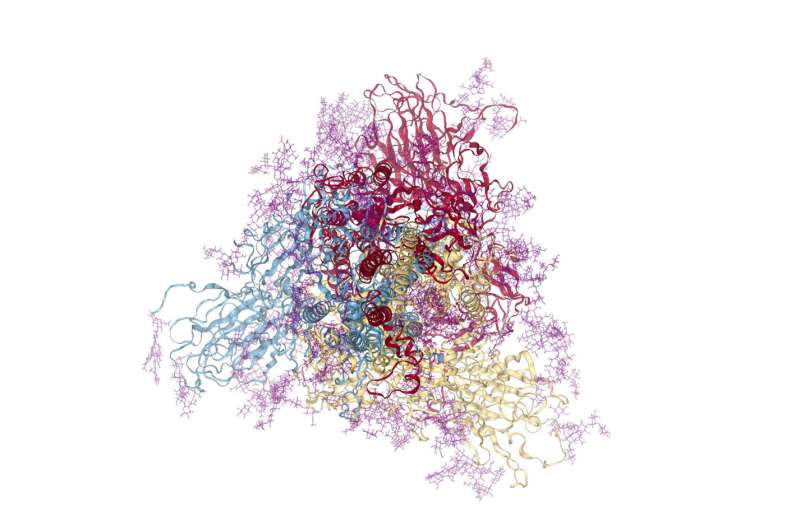
Designed to meet the urgent need for insights into the molecular intricacies of SARS-CoV-2 infection, BioExcel-CV19 is a repository for molecular dynamics (MD) simulations.
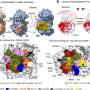
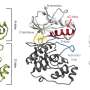
Researchers at IRB Barcelona, led by Dr. Modesto Orozco, have built this database, including key flexibility information essential for understanding the roles of key proteins in the virus, such as the Spike protein, Receptor Binding Domain (RBD), and RNA-dependent RNA polymerase (RdRp), providing over 10,000 trajectories.
"This database represents a shift in molecular dynamics databases, built to handle large systems and long trajectories while seamlessly integrating with modern MD simulations," explains Dr. Modesto Orozco, head of the Molecular Modeling and Bioinformatics lab at IRB Barcelona and University of Barcelona professor. "It brings together trajectories contributed by diverse research groups, showcasing a collaborative approach that drives scientific discoveries," he adds.
The article, published in Nucleic Acids Research, highlights the features of BioExcel-CV19 and underscores the importance of storing MD simulations. Stored simulations ensure result reproducibility and facilitate community-wide analysis. "Platforms like BioExcel-CV19 should become a standard, akin to the Protein Data Bank (PDB) in structural biology," states Dr. Adam Hospital, a Research Associate at IRB Barcelona, who has led this work together with Dr. Orozco.
BioExcel-CV19's impact extends to various scientific domains, including virology, genomics, structural and molecular biology, drug design, biomolecular simulation, and machine/deep learning. The open data provided becomes a valuable resource for training models, influencing fields that use data for various purposes.
In the field of biomolecular simulation, BioExcel-CV19 provides a wealth of data for refining algorithms and MD force fields. The potential for re-analysis and reuse of data opens avenues for new discoveries and meta-analyses, making BioExcel-CV19 a valuable asset for the scientific community.
Future efforts of the lab will focus on advancing BioExcel-CV19, ensuring it evolves with the ever-changing landscape of COVID-19 research. The database will continuously update with new simulations and user-suggested analyses, fostering ongoing collaborations and discoveries.
BioExcel-CV19 also serves as a prototype for an ambitious project called MDDB. Coordinated by IRB Barcelona, this European endeavor aims to design a European Repository for Biosimulation Data. With a consortium including the University of Oxford, Barcelona Supercomputing Center (BSC), CECAM, EMBL-EBI, KTH, and Nostrum Biodiscovery (NBD), MDDB represents a pioneering step towards federated and distributed infrastructures.
BioExcel-CV19 is a web-based portal that provides an accessible gateway for users to explore and query data interactively.
More information: Daniel Beltrán et al, A new paradigm for molecular dynamics databases: the COVID-19 database, the legacy of a titanic community effort, Nucleic Acids Research (2023). DOI: 10.1093/nar/gkad991
Journal information: Nucleic Acids Research