This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
proofread
GenoDrawing: Pioneering plant phenotyping with autoencoders and SNP markers
Advancements in whole-genome sequencing have revolutionized plant species characterization, providing a wealth of genotypic data for analysis. The combination of genomic selection and neural networks, especially deep learning and autoencoders, has emerged as a promising method for predicting complex traits from this data.
Despite the success in applications like plant phenotyping, challenges remain in accurately translating visual information from images into measurable data for genomic studies.
In November 2023, Plant Phenomics published a research article titled "GenoDrawing: An Autoencoder Framework for Image Prediction from SNP Markers."
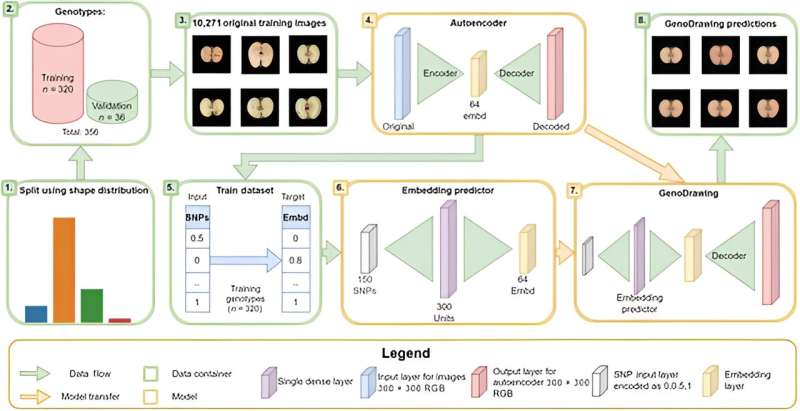
The study introduces an innovative approach using an autoencoder network and embedding predictor to simplify apple images into 64 dimensions and predict fruit shapes from molecular data (SNPs).
This method, known as GenoDrawing, involves training the autoencoder with a large dataset of apple images. The generated embeddings, along with SNP data, are then used to predict and reconstruct apple shapes.
The method showed that targeted SNPs (tSNPs) consistently outperformed randomly selected SNPs (rSNPs) in predicting image embeddings, resulting in more accurate fruit shape predictions.
The best models using tSNPs achieved lower mean absolute errors (MAEs) and produced distributions closer to the original data compared to rSNPs. Additionally, the tSNP-based version predicted a wider range of fruit shapes, demonstrating its effectiveness in capturing the diversity of apple phenotypes.
However, the study revealed limitations, including the model's inability to accurately capture certain fruit features and the influence of environmental factors on apple phenotypes.
Despite these challenges, the approach represents a significant advancement in genomic prediction, demonstrating the potential of merging image analysis with molecular data to comprehend complex traits in crops.
In summary, the findings suggest that selecting relevant SNPs is crucial for accurate predictions and that GenoDrawing can effectively learn to predict fruit shapes when given the appropriate markers.
This research lays the groundwork for future studies aiming to enhance the accuracy and applicability of genomic prediction models by incorporating image data and improving SNP selection strategies.
More information: Federico Jurado-Ruiz et al, GenoDrawing: An Autoencoder Framework for Image Prediction from SNP Markers, Plant Phenomics (2023). DOI: 10.34133/plantphenomics.0113
Provided by Plant Phenomics