This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Clustering algorithm helps scientists make sense of vast amounts of molecular data
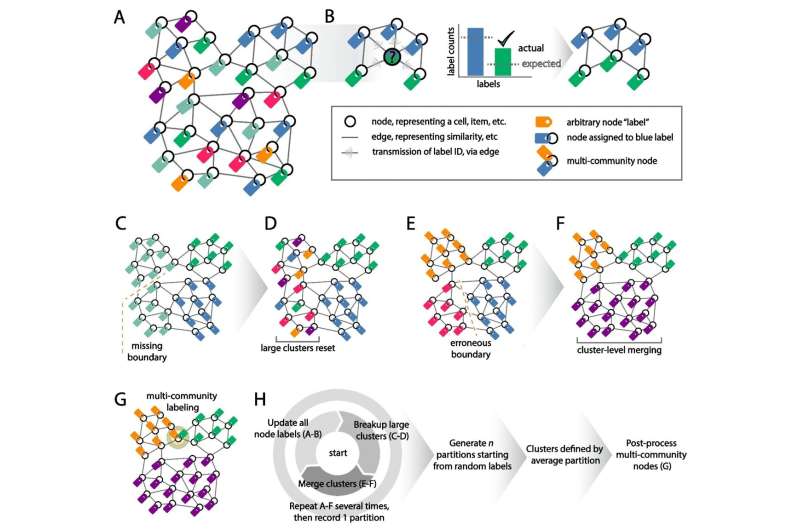
Thanks to technological advances, scientists have access to vast amounts of data, but in order to put it to work and draw conclusions, they need to be able to process it.
In research recently published in Genome Biology, Rensselaer Polytechnic Institute's Boleslaw Szymanski, Ph.D., Claire and Roland Schmitt Distinguished Professor of Computer Science and director of the Network Science and Technology Center, and team have found a method that effectively organizes and groups the data for a variety of applications. The process is referred to as clustering in machine learning.
Their clustering method, SpeakEasy2: Champagne, was tested alongside other algorithms to analyze its effectiveness in bulk gene expression, single-cell data, protein interaction networks, and large-scale human network data. Bulk gene expression tends to be tissue and disease-specific with implications on function and phenotype or how a genotype interacts with the environment.
Single-cell data is grouped according to a cell's distinctions. Protein binding is a core mechanism for signal propagation in cells, and identifying proteins that assemble into complexes is useful for defining functions within a cell.
The team's testing of SpeakEasy2: Champagne alongside other methods revealed that no single method is perfect for all situations, and the performance can vary. However, SpeakEasy2 performed well across different data types, suggesting that it's an effective way to organize molecular information.
"We tested to determine if the methods worked well even if the data included a lot of irrelevant information and also new, unseen data," said Szymanski. "We wanted to measure their reliability and performance in a number of ways, so we tested across a wide range of networks. SpeakEasy2: Champagne proved to have consistent and acceptable performance across diverse applications and metrics."
"Optimizing machine learning methods to integrate large amounts of noisy data effectively is critical to advancing science across many research fields," said Curt Breneman, Ph.D., dean of Rensselaer's School of Science. "Dr. Szymanski's work will allow new insights into cell function and gene expression and may illuminate new potential drug targets and their inhibitors to treat disease."
This work was done in collaboration with Chris Gaiteri, Ph.D., of Rush University Medical Center, and his team, resulting from a decade-long collaboration. Eight years ago, they collectively developed a novel clustering algorithm named SpeakEasy that, in light of vast new sources of biomedical data thanks to advances in computer science technologies, required more intelligent and faster software that would work for more diverse and greater amounts of biomedical data.
More information: Chris Gaiteri et al, Robust, scalable, and informative clustering for diverse biological networks, Genome Biology (2023). DOI: 10.1186/s13059-023-03062-0
Journal information: Genome Biology
Provided by Rensselaer Polytechnic Institute