This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Chromatin modifier-centered pathway points to higher crop yield
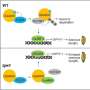
A team led by Prof. Song Xianjun from the Institute of Botany of the Chinese Academy of Sciences, while researching a ternary protein complex in rice nuclei that affects grain size, has shown that the transcription factor bZIP23—a protein that regulates the transcription of genetic DNA information into RNA and is part of the ternary complex—recruits the chromatin-modifying histone acetyltransferase HHC4 to specific promoters on the DNA.
Chromatin is the complex of DNA and proteins that makes up the genetic material in the nucleus of eukaryotic cells. A chromatin modifier is a protein or complex of proteins that chemically modifies the structure of chromatin.
Chromatin modifiers play a crucial role in regulating the expression of genes, which are segments of DNA strands, as well as in other chromatin-related processes. These modifiers mainly work by adding or subtracting chemical groups to histones, a type of protein within the chromatin, or to the DNA itself. In the scientific effort to manipulate the expression of plant genes, such as for grain size or drought resistance, etc., understanding the influence of chromatin modifiers is an essential avenue of research.
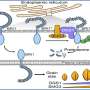
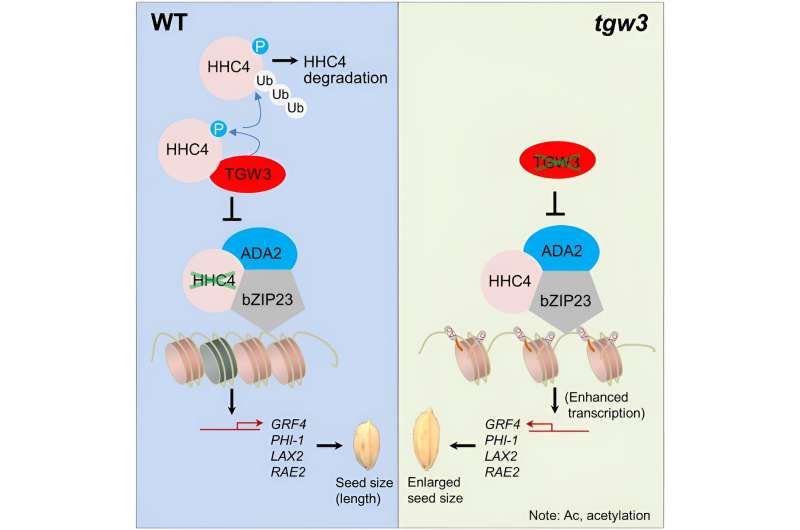
In their new study, the researchers discovered that HHC4 and the adaptor protein ADA2 additively enhance the bZIP23 transactivation of target genes. The researchers also showed that HHC4 is phosphorylated by the GSK3-like kinase TGW3, which triggers a series of negative influences on the capabilities of the ternary complex.
These findings, published in Developmental Cell, contribute to a deeper understanding of the epigenetic regulation of grain size.
A previous study by Prof. Song showed that GRAIN WEIGHT 6a (GW6a) encodes a newly identified histone acetyltransferase (OsglHAT1) that is a positive regulator of grain size and rice yield.
"At the beginning of this study, we identified five rice homologs of OsglHAT1 on chromosomes 2, 3, 4, and 7 (hence named HHCs) and sought to investigate whether these homologs also modulate grain size," said Dr. Shen Shao-Yan, first author of the study. "Interestingly, HHC4 regulates grain size but adopts a different cytological mechanism from GW6a."
Using a series of molecular biology techniques, including chromatin immunoprecipitation coupled with high-throughput sequencing (ChIP-seq) and immunoprecipitation followed by mass spectrometry (IP-MS), bZIP23 was shown to directly interact with HHC4.
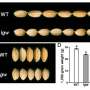
The bZIP23 gene has been characterized for its role in salinity and drought resistance as well as seed vigor in rice; however, whether it could regulate grain size was unknown. The researchers subsequently discovered that overexpression of bZIP23 significantly increased rice grain size, and bZIP23 and HHC4 co-targeted and synergistically activated the expression of several positive regulators of grain size.
Furthermore, the researchers found that HHC4, ADA2, and bZIP23 interact with each other, and the resulting ternary complex facilitates the additive enhancement of bZIP23 transactivation on target genes by ADA2 and HHC4.
Meanwhile, yeast two-hybrid (Y2H) screening revealed that the GSK3-like kinase protein TGW3 also interacts with HHC4. In addition, the researchers showed that HHC4 is directly phosphorylated by TGW3.
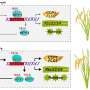
Mutation analysis suggested that two serine residues (S189 and S190) of HHC4 are major sites of phosphorylation by TGW3. Comparisons with mature grains of transgenic rice plants containing the mimicked phosphorylated and unphosphorylated versions of HHC4 suggested that phosphorylation is involved in grain size control.
Subsequent experimental data showed that phosphorylation exerts many negative influences, such as on the protein stability of HHC4, its interaction with bZIP23, and bZIP23's transactivation of target genes. In addition, the researchers showed that both HHC4 overexpression and TGW3 knockout substantially increased rice grain yield by up to 24% in field trials.
Overall, these findings uncover a previously unknown chromatin modifier-centered pathway for grain size regulation in rice and provide useful genetic resources for high-yield crop breeding programs.
More information: Shao-Yan Shen et al, Optimizing rice grain size by attenuating phosphorylation-triggered functional impairment of a chromatin modifier ternary complex, Developmental Cell (2024). DOI: 10.1016/j.devcel.2023.12.013
Journal information: Developmental Cell
Provided by Chinese Academy of Sciences