This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
A new approach can address antibiotic resistance to Mycobacterium abscessus
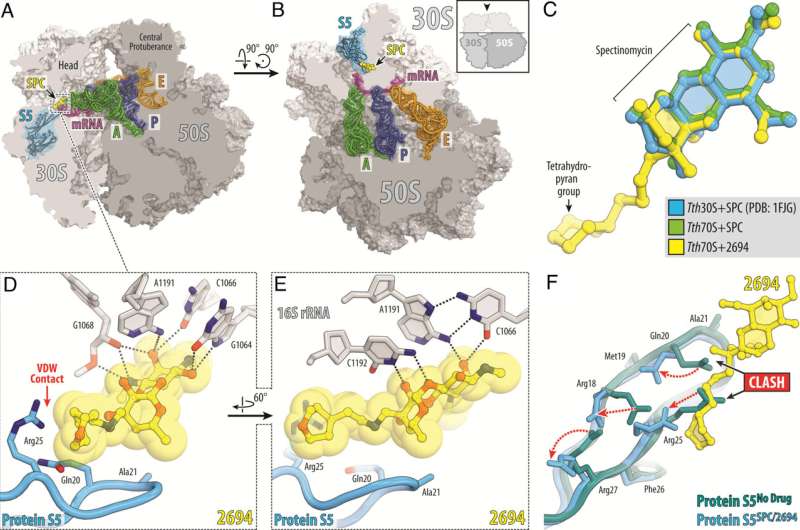
Scientists at St. Jude Children's Research Hospital are tackling Mycobacterium abscessus (Mab) antibiotic resistance. This naturally antibiotic-resistant pathogen is becoming more prevalent, highlighting the urgent need for novel therapeutics. To address this, the scientists designed new versions of the drug spectinomycin that overcome efflux, the main mechanism driving resistance. The work is published in Proceedings of the National Academy of Science.
Mab infections are increasingly found in health care settings. Such infections can be hazardous for patients with compromised lung function, such as in cystic fibrosis, or who are immunologically compromised, such as in childhood cancer. These infections are treated with long courses of antibiotics and can result in poor outcomes.
The emergence of Mab and other similar pathogens presents a growing and deeply concerning public health threat because there are few effective therapeutic options and a limited drug development pipeline.
"We chemists are in a race against the pathogens. We make stronger antibiotics, and the pathogens become more resistant," said corresponding author Richard Lee, Ph.D., St. Jude Department of Chemical Biology and Therapeutics.
Scientists at St. Jude modified the naturally occurring antibiotic spectinomycin to create analogs, comparable but structurally distinct N-ethylene linked aminomethyl spectinomycins (eAmSPCs). These synthetically created eAmSPCs are up to 64 times more potent against Mab than standard spectinomycin.
"By re-engineering the molecule through structure-based drug design, we and our collaborators have adapted the antibiotic to increase its activity," Lee added.
Overcoming efflux to make a more effective antibiotic
Through their work, the scientists unraveled the mechanism of action by which eAmSPCs are more effective: they circumvent efflux. Efflux is the process that cells use to get rid of a drug—imagine pumping water out of a flooded basement— and is a significant mechanism by which cells become resistant to therapy.
The N-ethylene linkage structure of the eAmSPCs plays a critical role in how the compounds avoid efflux, suggesting that longer linkages modify how the compound is pumped out of the cell. This ultimately shifts the balance toward higher concentrations of eAmSPC within the cell and thus enhances antimicrobial efficacy.
"Over the past two decades, we've seen a massive increase in the number of infections caused by non-tuberculous mycobacteria like Mab," said co-first author Gregory Phelps, PharmD, St. Jude Graduate School of Biomedical Sciences. "We had a place to start with this naturally occurring antibiotic, which, through modification, we've made much more efficacious against this clinically relevant pathogen."
The researchers also found that eAmSPCs work well with various classes of antibiotics used to treat Mab and retain their activity against other mycobacterial strains. This work demonstrates that eAmSPCs should be further studied and developed because once issues of tolerability and safety are addressed, these compounds could become next-generation therapeutics.
"It is challenging to attract pharmaceutical companies to develop new antibiotics for several economic reasons," said Phelps. "If we can boost the drug pipeline against this hard-to-treat bacteria, we can potentially make a difference for patients like the ones we have here at St. Jude who are increasingly faced with limited or no therapeutic options."
More information: Gregory A. Phelps et al, Development of 2nd generation aminomethyl spectinomycins that overcome native efflux in Mycobacterium abscessus, Proceedings of the National Academy of Sciences (2024). DOI: 10.1073/pnas.2314101120
Journal information: Proceedings of the National Academy of Sciences
Provided by St. Jude Children's Research Hospital