This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
A graphical user interface for analysis and design of protein–peptide interactions
Proteins function through interactions with other proteins or peptides, a complex process that is still not fully understood. Current research focuses on how amino acid residues interact in protein-protein or protein-peptide interactions to establish specific recognition. However, identifying key residues that are critical for binding affinity and specificity remains a challenge, and the design of specific protein-protein interaction interfaces remains a challenge for protein engineering.
In May 2023, BioDesign Research published a research article entitled "Atligator web: A graphical user interface for analysis and design of protein-peptide interactions."
In this study, the ATLIGATOR software and its web extension, ATLIGATOR web, were developed to enhance the analysis of protein interactions. ATLIGATOR identifies common interaction patterns among amino acids and stores them in a database, aiding in the understanding and engineering of new binding capabilities.
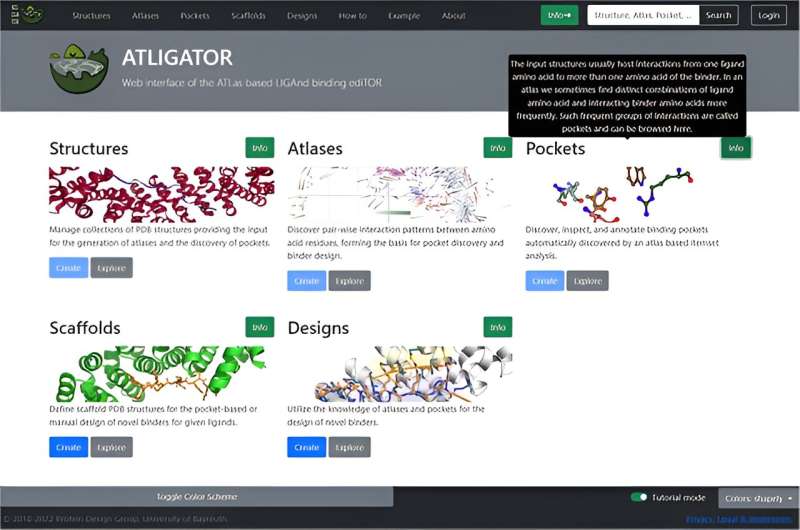
ATLIGATOR web, with its intuitive graphical user interface (GUI), expands the functionality of the original Python package, providing an accessible platform for users to explore pre-generated atlases and pocket collections and visualize interaction data. The results indicate that ATLIGATOR web improves user interaction through a structured interface comprising five main sections: Structures, Atlases, Pockets, Scaffolds, and Designs.
These sections are interconnected, facilitating easy navigation and detailed analysis. For example, the Atlases section derives pairwise interactions from protein structures in the Structures section, while the Pockets section groups recurring interaction motifs for visual analysis.
Additionally, the Scaffolds and Designs sections offer tools for practical application, allowing users to graft pockets onto protein structures and apply manual mutations for specific design tasks.
Overall, ATLIGATOR web not only makes the original ATLIGATOR tools more user-friendly but also introduces new functionalities like the advanced design tool for pocket grafting and rational design. This comprehensive and intuitive interface broadens the potential user base, encouraging the application of ATLIGATOR for detailed protein interaction analysis and design.
More information: Josef Paul Kynast et al, Atligator Web: A Graphical User Interface for Analysis and Design of Protein–Peptide Interactions, BioDesign Research (2023). DOI: 10.34133/bdr.0011
Provided by NanJing Agricultural University