This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
proofread
Study offers new method for predicting drug-protein binding affinity
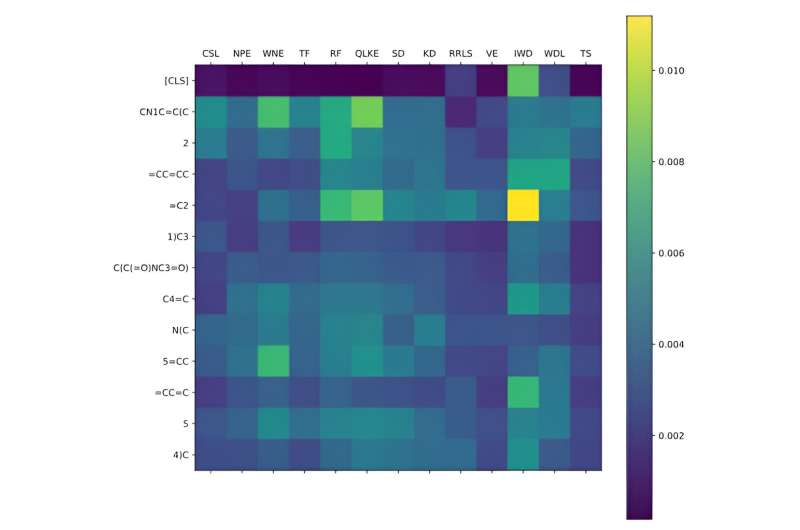
Accurately predicting the drug-protein interaction (DPI) is crucial in virtual drug screening. However, current methodologies tend to allocate equal weighting to amino acids and atoms in encoding protein and drug sequences, thereby neglecting the varying contributions from distinct motifs.
To tackle this issue, a group of researchers headed by Juan Liu have published their study in Frontiers of Computer Science.
Their research introduced a method, FragDPI, for the prediction of drug-protein binding affinity. This approach represents the initial endeavor to incorporate fragment coding and merge the sequence information of both drugs and proteins, hence preserving the primary features related to DPI interactions. Furthermore, this method employs transfer learning from significant DPI datasets to provide prospective DPI components.
-
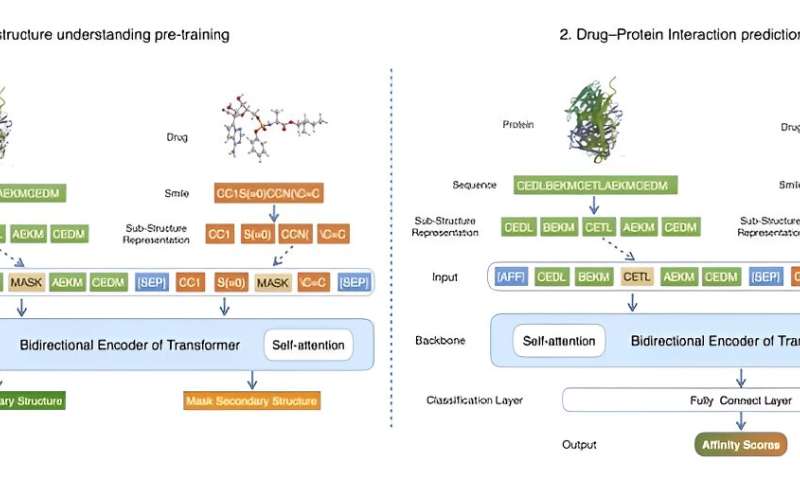
Credit: Frontiers of Computer Science (2022). DOI: 10.1007/s11704-022-2163-9 -
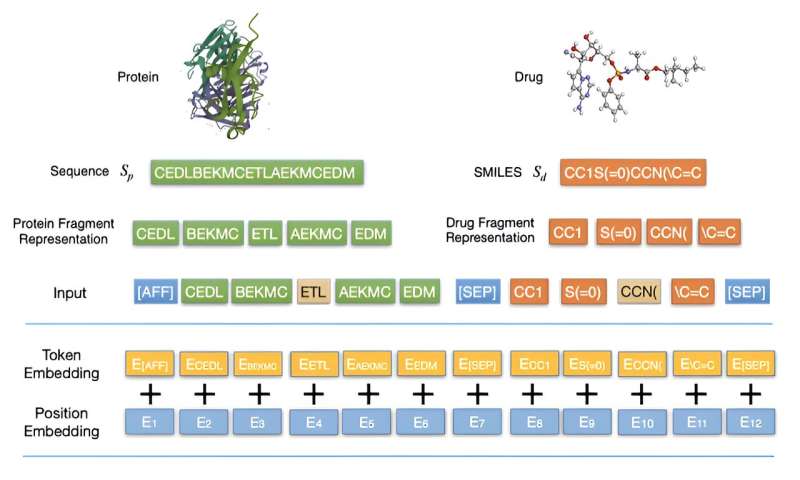
Credit: Frontiers of Computer Science (2022). DOI: 10.1007/s11704-022-2163-9 -
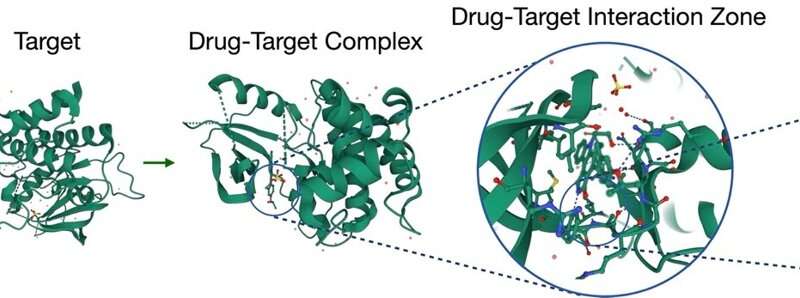
Credit: Frontiers of Computer Science (2022). DOI: 10.1007/s11704-022-2163-9
Experimental results demonstrate that the FragDPI model yields commendable outcomes compared to the baselines, including deep neural networks. Intriguingly, the model accurately identified the specific interaction parts of the DTI pairs, thereby aiding in discovering new potential DTI pairs.
FragDPI presents a novel approach for mining interacting fragments from DPI mechanism, thereby providing a fresh perspective towards drug discovery.
More information: Zhihui Yang et al, FragDPI: a novel drug-protein interaction prediction model based on fragment understanding and unified coding, Frontiers of Computer Science (2022). DOI: 10.1007/s11704-022-2163-9
Provided by Frontiers Journals