This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
proofread
A joint model for predicting circRNA-RBP binding sites based on deep learning
The interaction between circRNAs and RBPs is related to many diseases, especially cancers. Understanding the mechanism of the interaction and predicting their binding sites is critical.
New research published in Frontiers of Computer Science proposes a method to predict these binding sites using only sequence information of circRNAs.
The authors proposed a novel deep learning-based model named circ2CBA that considers the context information between sequence nucleotides of circRNAs and the important position weight information. The results of comparative and ablation experiments show that circ2CBA is effective.
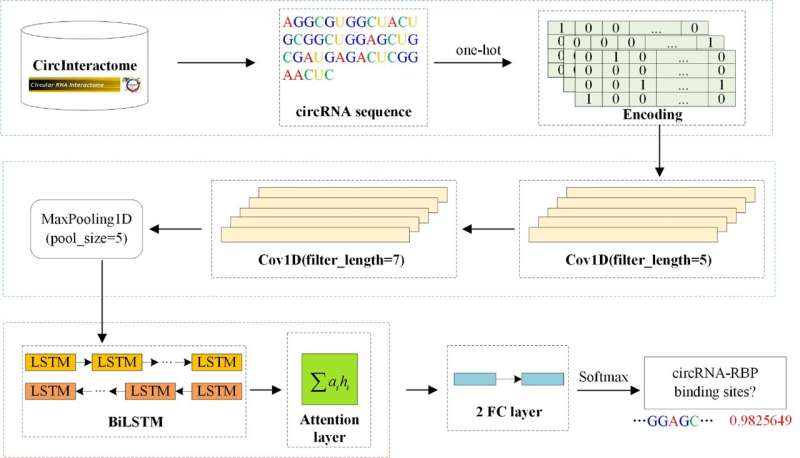
circ2CBA uses only sequence information of circRNAs to predict the binding sites between circRNAs and RBPs. The circRNA sequences were obtained from the CircInteractome database, and eight RBPs were selected to construct the data set.
The one-hot method is used to encode circRNA sequences, which are fed into subsequent models as input information. circ2CBA first utilizes two layers of CNN to extract the local features of circRNAs and to get a larger perception domain. To capture the context dependent information between sequence nucleotides, circ2CBA then introduces a BiLSTM network. Next, an attention layer is used to assign different weights to a feature matrix before feeding it to the two-layer fully connected layer.
Finally, the prediction result is obtained through a softmax function. Comparison experiments with other methods and ablation experiments were performed on the same dataset. In addition, a motif analysis explored the reason for the remarkable performance improvement of circ2CBA on some sub-datasets. The experimental results show that circ2CBA is an effective method to predict the binding sites of circRNA-RBP.
More information: Yajing Guo et al, circ2CBA: prediction of circRNA-RBP binding sites combining deep learning and attention mechanism, Frontiers of Computer Science (2022). DOI: 10.1007/s11704-022-2151-0
Provided by Frontiers Journals