This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
A bacterial toolkit for colonizing plants
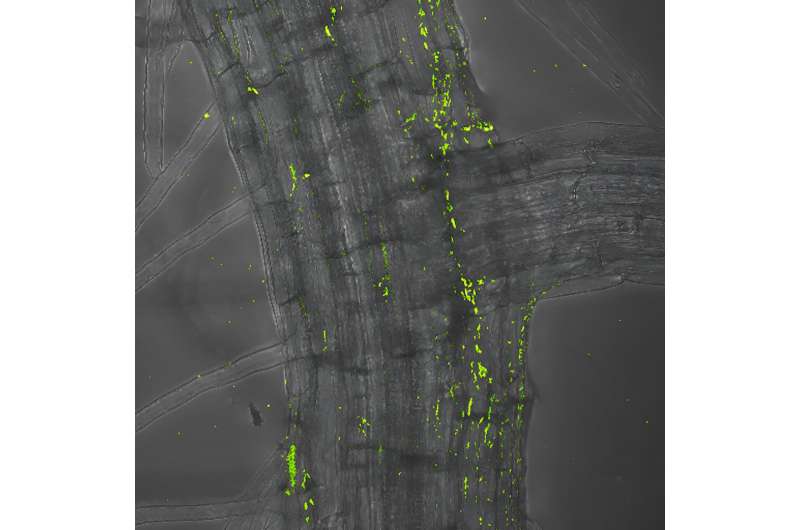
Using a novel experimental approach, Max Planck researchers have discovered a core set of genes required by commensal bacteria to colonize their plant hosts. The findings published in Nature Communications may have broad relevance for understanding how bacteria establish successful host–commensal relationships.
Plants are colonized by an enormous variety of microorganisms, including bacteria, archaea and fungi, that form complex communities, or microbiomes, on their roots and organs. Although invisible to the naked eye, the importance of these tiny inhabitants should not be underestimated. They play a crucial role in plant nutrition, influence the health of plants, strengthen their tolerance to stress factors such as drought and help defend against pathogens. Thus, harnessing the power of these microbial assemblages could contribute to more sustainable agriculture that is less reliant on fertilizers and pesticides.
Understanding how microorganisms colonize their hosts is a prerequisite for designing and applying microbiomes with beneficial functions, and, to this end, scientists often study how individual microorganisms interact with plants. However, researchers still don't have a good understanding of how multiple microbes concomitantly colonize and interact with plants for successful establishment of more complex host-commensal relationships.
This is frustrating, as information on how one microbe interacts with its host plant may not be representative of the complex reality observed in a microbial community context. The challenge has been essentially a technical one—how to precisely characterize the behavior of individual strains in a haystack of microbes and the plant itself.
To address the problem, Nathan Vannier and Stéphane Hacquard explored the establishment of individual microbes on plant roots in complex communities starting with microbe-free thale cress plants. Subsequently, they re-introduced a defined community of microbes, representing the diversity observed in the roots of plants in the wild, to investigate how these microbes colonize their host.
Knowing the identity of these bacteria and armed with reference sequences for their genetic material then allowed them to characterize what microbial genes were activated or repressed during plant colonization.
This analysis allowed them to identify numerous genes that were highly expressed in many different bacteria in roots and to select candidate genes potentially involved in host colonization. One regulates bacterial virulence and stress responses; another one is involved in transmembrane polymer transport and a set of genes that function together act as a phosphate sensor.
Mutating any three of these genes in bacteria hampered their ability to colonize roots, without affecting their growth in media. Thus, the authors' approach allowed them to identify a core set of genes required by many bacteria to persist on the roots of plants.
The strategy used by Hacquard and his team allowed them to understand both structural and functional organization of complex microbial communities that colonize plant roots. The genes identified may be broadly utilized by very diverse bacteria to colonize and persist on their hosts.
"Our results could potentially pave the way for engineering beneficial bacteria that can efficiently colonize host niches and promote host health. This has implications not only for sustainable agriculture but also for advancements in medical science," Stéphane Hacquard says.
More information: Nathan Vannier et al, Genome-resolved metatranscriptomics reveals conserved root colonization determinants in a synthetic microbiota, Nature Communications (2023). DOI: 10.1038/s41467-023-43688-z
Journal information: Nature Communications
Provided by Max Planck Society