Plant root–associated bacteria preferentially colonize their native host-plant roots

An international team of researchers from the Max Planck Institute for Plant Breeding Research and the University of Aarhus in Denmark have discovered that bacteria from the plant microbiota are adapted to their host species. In a newly published study, they show how root-associated bacteria have a competitive advantage when colonizing their native host, which allows them to invade an already established microbiota.
Plants, including crops such as rice and wheat, obtain their essential mineral nutrients and water through their roots, making them an important interface between plants and the soil environment. The roots of land plants associate with a wide range of microbes—including bacteria—that are recruited from the surrounding soil and assemble into structured communities known as the root microbiota. These microbial communities are sustained by the plant host, which provides them with nutrients, primarily in the form of organic carbon compounds secreted by the root. In turn, these commensal bacteria mediate multiple processes that are beneficial to their plant host, such as providing defense against pathogens, improving nutrient mobilization from the soil and positively impacting growth.
Given their importance for plant health, the study of the root microbiota has evolved into a promising research field that aims to understand how these interactions occur, and could eventually help increase the yield and resilience of crop plants. Although it is well known that plants secrete diverse small molecules into the soil via their roots that serve as chemoattractants for root colonization by a subset of soil-dwelling bacteria, the degree of active selection performed by the host and the extent to which root-associated microbial communities are adapted to specific plant species remain largely unknown. In a new study published in Nature Microbiology, a team of researchers from the Department of Plant-Microbe Interactions at the MPIPZ in Cologne, Germany, and Århus University in Denmark, aimed to gain a deeper understanding of these complex multi-species interactions.
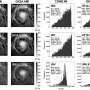
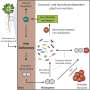
As a first step in this quest, they established a comprehensive collection of root-derived bacteria from the model legume Lotus japonicus, a small proportion of which are symbiotic bacteria that fix atmospheric nitrogen for plant growth. Together with an already established culture collection from roots of the model crucifer Arabidopsis thaliana, synthetic microbial communities (SynComs) were designed to explore the microbiota assembly of different plant species. Although the bacterial communities of the two plants were similar, the researchers observed a clear preference by these bacteria to colonize their native host. This preference was mediated by a higher competitiveness displayed by multiple bacterial species when colonizing their host of origin compared to those originally isolated from the other host.
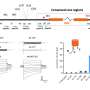
Strikingly, host preference was only observed in a community context, where different microbes compete among each other, but not when individual bacterial species were allowed to colonize the plant roots in the absence of competition. Analysis of gene expression of both plant species when interacting with different synthetic communities further showed that this process was at least in part driven by the host. Intriguingly, root colonization by native and non-native SynComs exhibited contrasting gene expression profiles for a number of well-known regulators of plant immunity. Based on this observation, the authors then hypothesized that native strains have a competitive advantage when colonizing the roots of their corresponding host plant via the formation of species-specific host niches. To test this hypothesis, the scientists performed a series of complex experiments, where SynComs from different host species were allowed to invade already established root-associated bacterial communities in host and non-host plants. Their results showed that native SynComs had a competitive advantage when invading an already established microbiota in their host plant, indicating that adaptation of commensal bacteria to their native plant species leads to increased invasiveness and persistence.
Kathrin Wippel, first author of the study, says: "We were amazed to learn that root colonization by native and non-native SynComs resulted in differential transcriptional reprogramming of plant roots, possibly contributing to the formation of specific root niches for native commensal bacteria. These findings indicate that diverse soil-dwelling bacteria associate with and prefer a specific host plant, similar to pathogens or beneficial symbionts of plants."
These findings could have a meaningful impact on agriculture, as they highlight the importance of competitiveness between different bacteria and the impact of host preference for successful root colonization. Probiotic inoculants tailored to specific crop plants with an enhanced capacity to invade and persist in standing microbial communities could help overcome the variation in efficacy of currently used biologicals in agriculture.
More information: Host preference and invasiveness of commensal bacteria in the Lotus and Arabidopsis root microbiota, Nature Microbiology (2021). DOI: 10.1038/s41564-021-00941-9 , www.nature.com/articles/s41564-021-00941-9
Journal information: Nature Microbiology
Provided by Max Planck Society