This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Structurally editable proximal cofactor-like module helps to construct artificial dual-center peroxygenase
Cytochrome P450 monooxygenases are widely involved in the synthesis and metabolism of endogenous and exogenous substances in living organisms. The catalytic efficiency of cytochrome P450 monooxygenase relies on the coenzyme NAD(P)H and reducing chaperone proteins.
The strategy based on dual-functional small molecules (DFSMs) can covert P450 monooxygenase into peroxygenase, which avoids utilizing the expensive coenzyme and complicate chaperone proteins. However, excess DFSMs are required owing to their low binding affinity for P450, limiting its practical application.
To solve this issue, researchers from the Qingdao Institute of Bioenergy and Bioprocess Technology (QIBEBT) of the Chinese Academy of Sciences (CAS) have developed a structurally editable proximal cofactor-like module for constructing an artificial dual-center peroxygenase.
The study was published in Angewandte Chemie International Edition on Oct. 27.
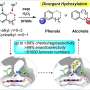
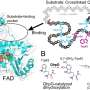
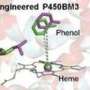
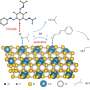
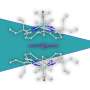
The researchers constructed an artificial dual-center peroxygenase by anchoring an editable organic cofactor to the proximal position of the heme center of P450BM3 as a co-catalytic center. The co-crystal structure of P450BM3 in complex with the novel artificial cofactor clearly revealed a precatalytic state in which the cofactor participated in H2O2 activation, thus facilitating peroxygenase activity.
Compared with previous DFSMs, the novel artificial cofactors could form more hydrogen bonds and hydrophobic interactions with the enzyme, suggesting a much higher binding affinity. Furthermore, the dissociation constants (Kd) of novel cofactors were accurately determined through titrations. The Kd values of some artificial cofactor were increased by three orders of magnitude and comparable to the binding efficiency of natural enzyme cofactors.
Enzyme activity measurements showed that even with the addition of only a small amount of new artificial cofactors (twice the enzyme amount), the system still exhibited high catalytic activity for typical P450 enzyme oxidation reactions such as olefin epoxidation, hydroxylation of sp3-carbons, and thioether oxidation.
Moreover, the researchers found that different catalytic groups, such as imidazole, pyridine or amine groups, had divergent catalytic activity and selectivity for substrates. Therefore, different types of new cofactors would be selected based on the properties of substrates to achieve the optimal catalytic effect in future applications.
More information: Xiangquan Qin et al, Cover Picture: Anchoring a Structurally Editable Proximal Cofactor‐like Module to Construct an Artificial Dual‐center Peroxygenase, Angewandte Chemie International Edition (2023). DOI: 10.1002/anie.202315458
Provided by Chinese Academy of Sciences