This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
proofread
Exploring spatial transcriptomics in biomedical research
Spatial transcriptomics (ST) emerges as a pivotal technique for cataloging gene expression across tissue sections while retaining crucial locational data.
Traditional methodologies, encompassing bulk or single-cell RNA sequencing, are incapable of simultaneously capturing both gene expression profiles and the spatial locations of cells.
ST distinguishes itself by revealing the concealed spectrums of cellular heterogeneity, organizational patterns, and molecular communications foundational to tissue structure and dynamics. Moreover, ST has the capacity to coalesce with other 'omics' strategies, thus offering a synthesized and expansive perspective of biological entities across various levels of detail.
With ST's inception, there has been the introduction of novel methods that enhance throughput and resolution, promising substantial advances in the elucidation of biological intricacies.
In research, published in Molecular Biomedicine, researchers systematically categorize and contrast various ST techniques according to their foundational principles and procedural intricacies, offering a thorough assessment of the array of ST approaches. This evaluation highlights the fundamental trade-off that must be navigated between obtaining high-resolution data and ensuring a high-throughput workflow.
Recent innovations, such as Stereo-seq, demonstrate significant strides towards resolving this dichotomy; however, the quest to achieve the gold standard—meticulous localization of each transcript at the cellular level with exacting precision—persists.
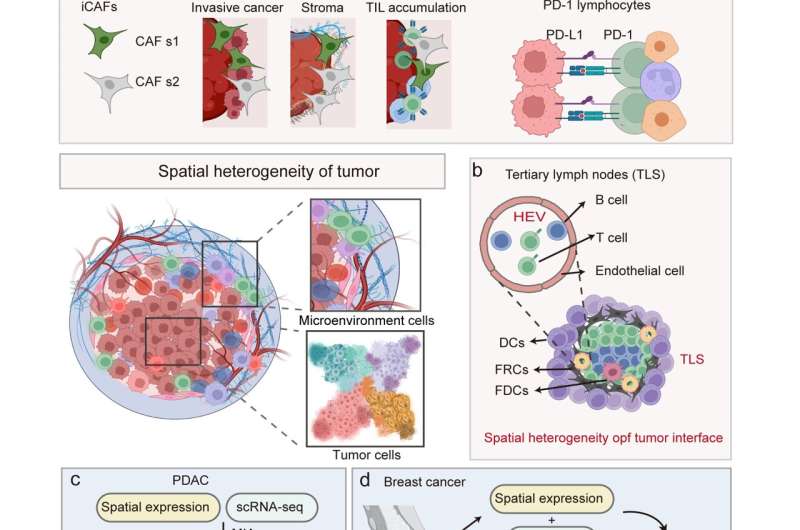
This review elucidates the utility of spatial transcriptomics (ST) across a breadth of biomedical research fields, encompassing developmental biology, neuroscience, immunology, and oncology. ST has been instrumental in delineating the complex cellular dynamics within tissue architectures, their mutual interactions, and their pivotal functions in sophisticated biological processes, thus yielding transformative insights into an array of pathological conditions from cancers to neurodegenerative diseases.
They examine ST's proficiency in demystifying the multilayered complexities of the tumor microenvironment, confronting enduring challenges such as intratumoral heterogeneity that have traditionally thwarted efficacious oncological interventions.
The review discusses the current limitations and challenges in the field. Such as the diverse range of methodologies that leads to a multitude of file formats and data structures leading data and protocol sharing more difficult. Furthermore, there exists a significant hurdle in harmonizing spatially-resolved data across both intra-omics and cross-omics layers.
Finally, this review contemplates prospective trajectories for the advancement of spatial transcriptomics, accentuating the necessity to enhance spatial resolution, expand gene coverage, and simplify the operational complexity of current methodologies. There exists an urgent need for the development of new computational tools designed to enhance the processing, and integration.
The authors summarize the recent advancement of ST in historical, technical, and application contexts. The review serves as a valuable reference, guiding researchers in selecting the most apt spatial transcriptomics methodologies tailored to their unique biological inquiries and experimental conditions.
More information: Ran Zhou et al, Spatial transcriptomics in development and disease, Molecular Biomedicine (2023). DOI: 10.1186/s43556-023-00144-0
Provided by Sichuan International Medical Exchange and Promotion Association