This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Research on beer fermentation yeast reveals unexpected evolutionary process
Researchers at the University of Wisconsin–Madison made an unexpected discovery while studying a strain of yeast closely related to the kind used to ferment beer—they observed the yeast left half its genetic material behind while evolving.
Their study, published this week in the journal PLOS Biology, started with the straightforward goal of investigating a strain of the wild yeast Saccharomyces eubayanus, a wild parent of the hybrid yeast strains used in lager-style beer fermentation.
A key trait of yeasts used in beer production is the ability to break down maltose, the most abundant sugar in the material used to brew beer. The team chose a yeast strain that had evolutionarily lost this ability despite possessing many of the genes required for maltose metabolism. After running evolution experiments growing the yeast on maltose and selecting decedents that thrived, the lab's S. eubayanus yeast reacquired the ability to eat maltose.
However, it was not immediately clear how the yeast changed.
"We kind of banged our heads against the wall because we couldn't really make sense of figuring out the underlying genetic changes of these adaptively evolved isolates that had improved their maltose metabolism," says John Crandall, first author of the paper and a doctoral student in the lab of Chris Todd Hittinger, UW–Madison genetics professor.
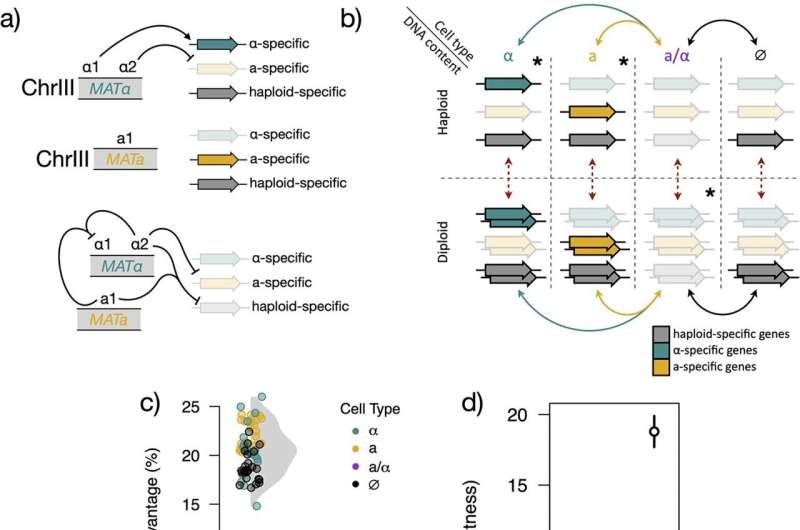
There was not just a simple mutation in any specific gene—the change was much more dramatic: the yeast had lost half of its genetic material during the experiment. More specifically, all chromosomes lost their pair, turning from diploids (with two copies of each of its chromosomes, like humans have) to haploids (with a single copy of each chromosome, like our gametes, or eggs and sperm).
The number of sets of chromosomes in organisms, known as ploidy, is one of the most fundamental defining aspects of organismal biology.
"It defines the structure and content of the genome, which then has all sorts of implications for life cycle and evolutionary trajectories," Crandall says.
Ploidy changes in yeasts have been documented before, but usually haploids become diploids and not the other way around. The ploidy change in yeasts also frequently causes a change in cell type, which directly affects sexual reproduction. But here, it was affecting sugar metabolism.
"So there were lots of different factors to sort of disentangle between the absolute ploidy, the cell type, how those different things contributed to fitness [growth and reproduction]. And that interested us, because there are some different theoretical frameworks about why absolute ploidy itself might impact fitness," explains Crandall. "But there wasn't a good model for why cell type should affect fitness in this condition, which is also why it's particularly surprising that it did."
The new study is perhaps the clearest mechanistic insight into a ploidy change that directly affects growth and metabolism. This insight could allow for multiple applications in diverse fields, such as medicine, biofuel production, and many products and industries that involve fungi.
"In lots of pathogenic and clinically or industrially relevant species that people are interested in, cell type or ploidy might represent an underexplored dimension of trait variation," Crandall says.
More information: Johnathan G. Crandall et al, Ploidy evolution in a wild yeast is linked to an interaction between cell type and metabolism, PLOS Biology (2023). DOI: 10.1371/journal.pbio.3001909
Journal information: PLoS Biology
Provided by University of Wisconsin-Madison