This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
How bacteria recognize viral invasion and activate immune defenses
There's no organism on Earth that lives free of threat—including bacteria. Predatory viruses known as phages are among their most dire foes, infiltrating their cells to replicate and take over. Bacteria have evolved an array of strategies to counter these infections, but how they first spot an invader in their midst has long been a mystery.
Now researchers in the Laboratory of Bacteriology at The Rockefeller University have discovered that bacteria sense phages via a defensive response called CBASS that detects viral RNA—findings that one day may help counter the threat of antibiotic resistance. They published the results in Nature.
"How CBASS is activated by phage infection has been a big unknown in our field for many years," says Luciano Marraffini, head of the lab. "Until now, no one has understood what triggers the bacteria to initiate the CBASS immune response."
Kin across distant domains
Some core immune functions are shared across distantly related domains of life, from eukaryotes (organisms with a membrane-bound nucleus) like mammals, plants, and fungi to prokaryotes (those without such membranes) like bacteria and archaea. These immune responses must've evolved early in the existence of life.
One conserved characteristic is a viral sensing mechanism that relies on a specialized enzyme known as a cyclase. In animals, it's called cGAS (cyclic GMP-AMP synthase). In bacteria, cGAS-like cyclases are central components of the CBASS (cyclic oligonucleotide-based antiphage signaling system) immune response. Both were only discovered in the past decade.
"CBASS cyclases are thought to be ancient ancestors of cGAS," says co-first author Dalton Banh, an M.D.-Ph.D student in Marraffini's lab.
But there are some differences. In an infected animal, cGAS senses viral DNA in the cytoplasm, the gelatinous liquid in a cell that surrounds the nucleus; in an uninfected organism, DNA is confined within the nucleus. Its presence elsewhere signals that something is amiss.
However, because bacteria lack nuclei, they must take another approach. If CBASS reacted to the mere presence of DNA, it would result in rampant autoimmunity, or the bacterium attacking itself, Banh says.
"That was the conundrum," he says. "CBASS cyclases look a lot like cGAS, so they have to be sensing something. But what, exactly?"
RNA seeker
To find out, the researchers and their collaborating partners in Sean Brady's Laboratory of Genetically Encoded Small Molecules focused on the CBASS system in Staphylococcus schleiferi, a bacterium commonly found in the mouths of dogs, cats, and other animals that on rare occasions has jumped to humans.
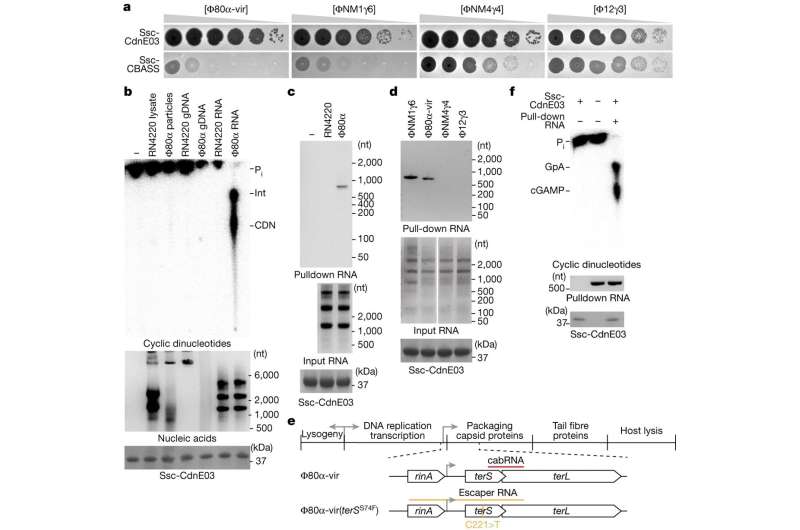
Marraffini is a pioneer of the study of bacterial defense systems, primarily CRISPR-Cas; because his lab has used a variety of Staphylococcus strains in this work over the years, the team has a lot of Staph phages on hand. Banh screened them all for their ability to be inhibited by CBASS. He homed in on a set of phages that were spotted by the defense system. "This led us to hypothesize that these sensitive phages produced something during infection that triggered activation of CBASS," Banh says.
Next, co-first author Cameron Roberts, a Ph.D. student in the lab, meticulously tested a variety of molecules produced by either the bacterium or the virus, including DNA, RNA, and proteins.
The experiment revealed that only RNA produced during phage infection was able to trigger an immune response. "It was very clearly viral RNA that was generated during infection," says Roberts. "So instead of sensing a DNA mislocalization, like cGAS does, CBASS senses a specific RNA structure. This specificity is amazing."
They coined the newly identified, hairpin-shaped molecule cabRNA (pronounced "cab-R-N-A" or alternatively, "cabernet"), for CBASS-activating bacteriophage RNA. The molecule binds to a surface of the cyclase, triggering the production of a messenger molecule called cGAMP that activates the CBASS immune response.
"It was a very simple and elegant experiment, and it gave us the key finding," Marraffini says.
Here, too, there are parallels to how the analogous system operates in humans. After detecting viral DNA, cGAS also triggers the production of cGAMP, which induces the immune system to produce Type I interferons. That antiviral signaling pathway is known as cGAS-STING.
Future possibilities
In future research, Roberts will continue to analyze cabRNA for its characteristics. "Two big questions are how and why the phage generates cabRNA—what is its role?" she says. "The details of how cabRNA interacts with the CBASS enzyme is also unclear. So solving a structure of the enzyme as it's bound to the cabRNA would be a huge feat."
The phages that don't trigger a CBASS response might potentially be useful one day in combating antimicrobial resistant bacteria. "Right now, we don't have the knowledge to predict which phages have the cabRNA and which phages don't," Marraffini says, "but if we could do that, we could potentially use those phages to attack bacteria, because they've figured out how to slip by this sensing mechanism."
More information: Luciano Marraffini, Bacterial cGAS senses a viral RNA to initiate immunity, Nature (2023). DOI: 10.1038/s41586-023-06743-9. www.nature.com/articles/s41586-023-06743-9
Journal information: Nature
Provided by Rockefeller University