This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Reassembly of parallel trimolecular G-quadruplex via novel Hoogsteen strand displacement reaction
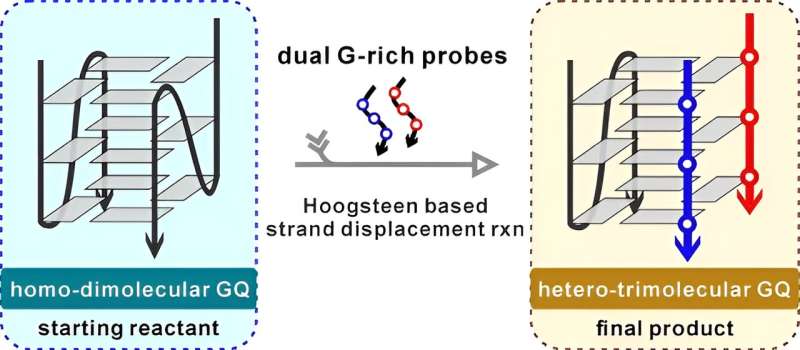
Using solution-state Nuclear Magnetic Resonance (NMR) technology, a group of scientists led by Prof. Zhang Na from Hefei Institutes of Physical Science (HFIPS) of Chinese Academy of Sciences (CAS) recently discovered a novel reassembly process for G-quadruplexes (GQs) through a new type of reaction called Hoogsteen pairing-based Strand Displacement Reaction (Hoogsteen SDR).
Conventional Watson-Crick SDR involves the displacement of one strand from a double-stranded DNA or RNA helix by a single-stranded homologous invading strand, completely based on the principle of the Watson–Crick base-pairing rule. GQs, which are noncanonical secondary structures formed by guanine-rich nucleic acid fragments, can exist in different forms like monomeric or intermolecular assemblies. Well-folded GQs are highly stable and resistant to SDR with other guanine-rich strands.
"Our work is the first to describe the spontaneous reassembly of G-quadruplex via Hoogsteen pairing-based SDR and present a novel NMR solution structure of heteromeric tri-GQ with a unique mode of two probes vs. one target," said Professor Zhang Na.
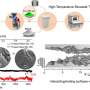
In this research, now published in the Journal of the American Chemical Society, the team focused on a specific target sequence of Tub10 d(CAGGGAGGGT), which was a DNA fragment from the G-rich promoter region of the human β2-tubulin gene.
In a K+ solution, the Tub10 sequence self-folds into a parallel homomeric dimolecular GQ (di-GQ) with high thermal stability. The scientists utilized Hoogsteen pairing-based SDR and introduced a pair of short G-rich probes (P1 d(TGGGA)) to invade the Tub10 GQ. Through this process, they were able to reassemble the starting di-GQ into a novel parallel heteromeric trimolecular GQ (tri-GQ) of Tub10/2P1.
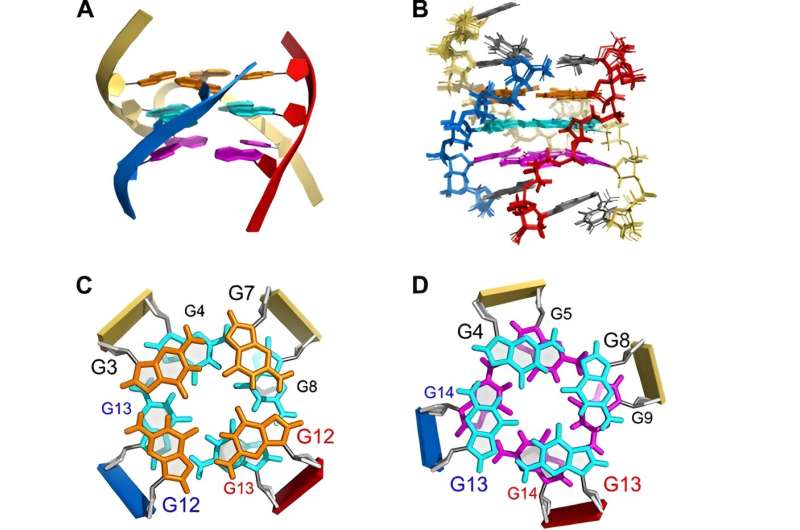
This research not only provided the first NMR solution structure of a discrete heteromeric tri-GQ but also revealed a unique mode of recognition between two probes and one target among G-rich DNA fragments. The short G-rich probe P1 demonstrated higher specificity for GQ targets compared to traditional antisense probes. Furthermore, P1 served as a model system by effectively capturing the G-rich target Tub10 from a Watson-Crick duplex formed when Tub10 hybridized with its complementary strand.
These findings open up new possibilities for the reassembly of GQs and offered insights into the interaction between G-rich DNA fragments, according to the team.
More information: Wenxuan Hu et al, Conversion to Trimolecular G-Quadruplex by Spontaneous Hoogsteen Pairing-Based Strand Displacement Reaction between Bimolecular G-Quadruplex and Double G-Rich Probes, Journal of the American Chemical Society (2023). DOI: 10.1021/jacs.3c05617
Journal information: Journal of the American Chemical Society
Provided by Chinese Academy of Sciences