This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Global phosphoproteomics reveal the diverse roles of casein kinase 1 in plant development
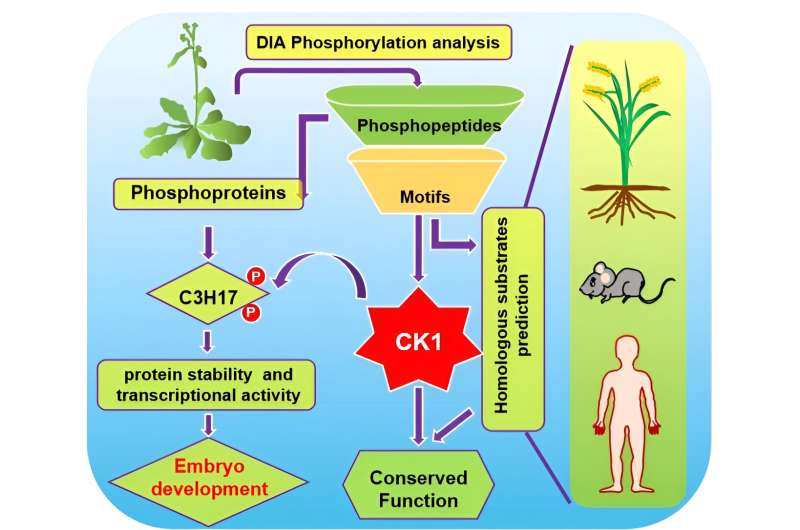
Protein phosphorylation is a major posttranslational modification carried out by protein kinases that constitute an integral part of complex signaling networks in eukaryotes. Casein kinase 1 (CK1) is a conserved serine/threonine protein kinase in eukaryotes and plays pivotal roles in both plants and mammals through phosphorylating various substrates.
Despite the emerging importance of CK1, the systematic disclosure of its function is still limited by the precise identification of endogenous substrates. High-throughput analysis of CK1-mediated phosphorylation events will facilitate the characterization of endogenous substrate and functional clarification of CK1.
Phosphoproteomics empowers a more effective profiling of phosphoproteome in vivo. A recent technological advancement in sampling, data-independent acquisition (DIA), makes it possible to identify and collect phosphopeptides with higher identification range and throughput, significantly improving the sensitivity and accuracy for LC-MS results.
In this study, Qu et al. performed quantitative DIA-based phosphoproteomics to study site-specific phosphorylation events mediated by CK1/AELs in Arabidopsis. They identified 3985 CK1-dependent phosphopeptides and defined 1,032 phosphoproteins through detecting and analyzing seedlings overexpressing individual AEL genes or lacking AEL functions.
Interestingly, a CCCH-type zinc finger transcription factor, C3H17, was identified in the embryonic development related substrate enrichment. Further biochemical and genetic studies proved that AELs promote protein stability and transcriptional activity of C3H17 through phosphorylation to regulate plant embryo development, providing informative clues for complex regulation of plant reproductive development.
Subsequently, four new CK1s substrate motifs were enriched among CK1-upregulated phosphopeptides and verified. Searching Arabidopsis protein database with these motifs yield multiple unreported biological processes which CK1-mediated phosphorylation may involve, as well as new putative substrates or novel mechanisms in known biological processes.
As CK1 is highly conserved across eukaryotes, they searched the rice, mouse, and human protein databases with the verified motifs and enriched 2,099 animal and plant-conserved biological processes, which greatly expands the potential substrate range and possible mechanism of CK1, and facilitates the functional studies and mechanism elaboration of CK1. In a wider scope, these data provide insights into the conserved function of CK1 in plants and animals.
This study obtained CK1-dependent phosphopeptides and phosphorylated proteins as well as substrate motifs of CK1 by phosphoproteomics analysis. Functional analysis of substrate C3H17 elucidated the regulatory mechanism of CK1-mediated phosphorylation in embryonic development.
These conserved functions and newly predicted substrates will help to study the essential and general roles of CK1s across species, opening up new avenues for the study of gene function in the context of basic research and for translational medicine applications.
The study is published in the journal Science Bulletin.
More information: Li Qu et al, Data-independent acquisition-based global phosphoproteomics reveal the diverse roles of casein kinase 1 in plant development, Science Bulletin (2023). DOI: 10.1016/j.scib.2023.08.017
Provided by Science China Press