This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
A new way to study molecular drivers of cancer
A clearer understanding about the markers and drivers of cancer cell proliferation has emerged from research that identifies new opportunities to overcome convergence with complex enzymes, known as kinases. The work paves the way for new approaches to study the molecular drivers of disease states such as cancer.
The research—"CDK6 activity in a recurring convergent kinase network motif," by Christina Gangemi, Rahkesh Sabapathy and Harald Janovjak—has been published in the Federation of American Societies for Experimental Biology Journal (The FASEB Journal).
Kinases are a specific family of proteins that add phosphates to other molecules—a process called phosphorylation, which can change the function of their substrates (target proteins). In humans, more than 500 kinases phosphorylate approximately 15% of all proteins. However, more than one kinase can phosphorylate the same substrate, and this can occur at the same or different sites. This is known as convergence, and can often make it difficult to study a specific kinase or substrate, as the activity of multiple kinases can hamper analysis.
Understanding the complex kinase network is important, as dysregulation of these proteins can drive disease, such as the survival and spread of cancer cells or their resistance to therapeutics.
While most kinase research has tended to focus on characterizing phosphorylation networks between kinases and their substrates, researchers in the Janovjak Lab at Flinders University's College of Medicine and Public Health have taken a different tack by analyzing how common convergence is across all human kinases, and using these insights to dissect it experimentally.
One example is the closely related cyclin-dependent kinases 4 and 6 (CDK4/6).
"Many kinases, particularly cell cycle proteins such as CDK4/6, are drivers of cancer cell proliferation and drug resistance. Our aim was to find new avenues to study them more effectively, as well as analyze and dissect convergence across all human kinases," says Christina Gangemi, a research associate at the Flinders Health and Medical Research Institute.
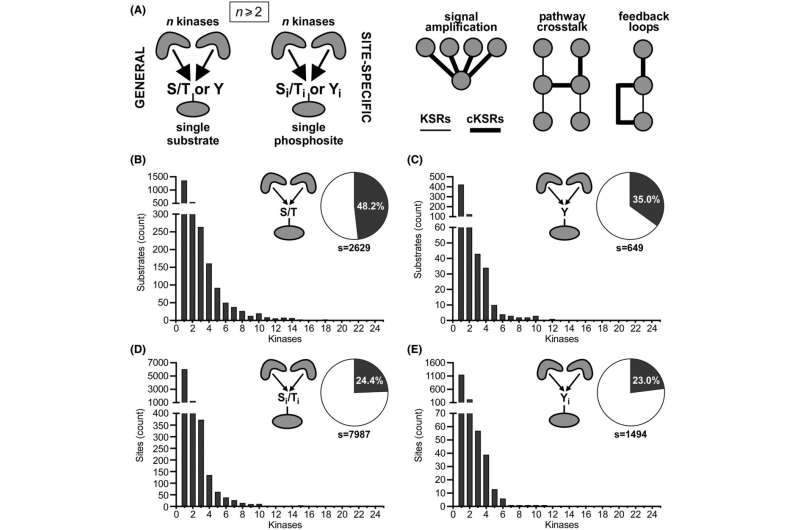
The researchers first used computational approaches to quantify and map convergent kinase-substrate relationships (cKSRs). Results revealed that cKSRs are common and involve more than 80% of all human kinases and more than 24% of all substrates. Convergence is also widespread when more than one kinase is phosphorylating the same site on a substrate (site-specific cKSRs).
The research also identified signaling pathways where reciprocal phosphorylation loops occur, when the substrate of a kinase can phosphorylate that kinase. Interestingly, around half of these interactions have not been previously described.
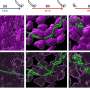
Co-expression analysis revealed that cKSRs are often expressed in the same cell type. For instance, convergent serine/threonine kinases are co-expressed in more than 70% of the study's analyzed human cell lines.
The researchers took these new insights to dissect the prototypical convergent kinase pair, CDK4/6, which are closely related family members that phosphorylate the retinoblastoma protein (RB) and share overlapping phosphorylation sites.
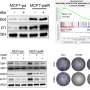
In most contexts, the presence of either CDK4 or CDK6 is sufficient to phosphorylate RB to levels that prohibit analysis of the activity of the other kinase. Also, there are CDK4/6 inhibitors, but not any specific to CDK6 or CDK4 alone, which makes it technically difficult to study them individually. The researchers first over-expressed CDK6 in a breast cancer cell line to see if its effect on RB phosphorylation could be detected.
"We found that simple over-expression of CDK6 in these cells is not sufficient to detect its activity, as RB is already highly phosphorylated by CDK4, so we had to take an alternate approach," says Ms. Gangemi.
The addition of a CDK4/6 inhibitor that has previously been shown to drive CDK6-mediated inhibitor resistance in breast cancer cells was employed. CDK6 over-expression combined with inhibitor treatment resulted in detection of CDK6-dependent RB phosphorylation.
The assay can be used for a range of purposes such as understanding the effect of protein modulators on CDK6 function, testing engineered versions of the kinase, or studying the effects of mutations that may be involved in driving disease.
"Along with developing a new assay to study CDK6, the computational analysis we performed has the potential to dissect other cKSRs. This work provides new opportunities to overcome convergence with kinases experimentally and could move us towards a deeper understanding of kinase networks and functions," says Gangemi.
More information: Christina G Gangemi et al, CDK6 activity in a recurring convergent kinase network motif, The FASEB Journal (2023). DOI: 10.1096/fj.202201344R
Journal information: FASEB Journal
Provided by Flinders University