This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Split gene-editing tool offers greater precision
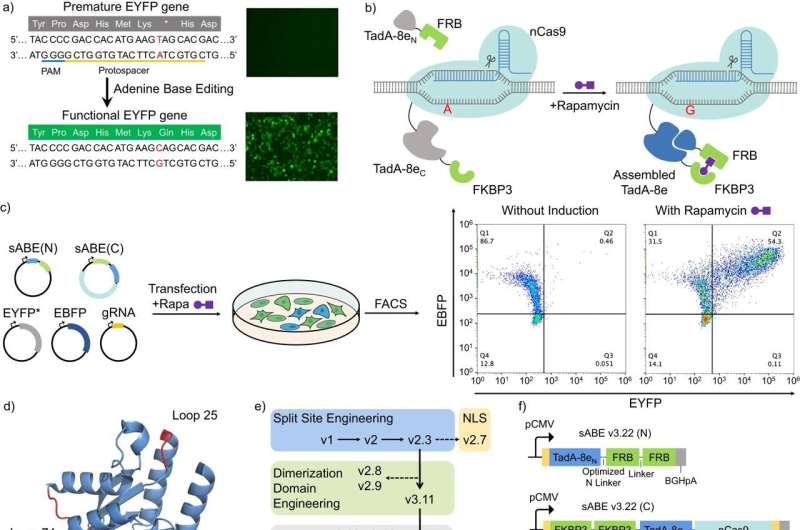
To make a gene-editing tool more precise and easier to control, Rice University engineers split it into two pieces that only come back together when a third small molecule is added.
Researchers in the lab of chemical and biomolecular engineer Xue Sherry Gao created a CRISPR-based gene editor designed to target adenine—one of the four main DNA building blocks—that remains inactive when disassembled but kicks into gear once the binding molecule is added.
Compared to the intact original, the split editor is more precise and stays active for a narrower window of time, which is important for avoiding off-target edits. Moreover, the activating small molecule used to bind the two pieces of the tool together is already being used as an anticancer and immunosuppressive drug.
According to a study published in Nature Communications, the tool developed by Gao and collaborators performed well both in human cell cultures and in living mice, where it accurately edited a single base pair on a target gene. Given that single base-pair mutations—also known as point mutations—are responsible for thousands of diseases, the split editor could have broad therapeutic applications.
"This tool has the potential to correct nearly half of the disease-causing point mutations in our genome," said Hongzhi Zeng, the lead author of the study and a graduate student in the Gao lab. "However, current adenine base editors are in a constant 'on' state, which could lead to unwanted genome changes alongside the desired correction in the host genome.
"Our team set out to create a much improved version that can be turned on or off as needed, providing an unparalleled level of safety and accuracy."
To install an "on/off" switch, the researchers broke the adenine base editor into two separate proteins that remain inactive until sirolimus (previously known as rapamycin) is added—a molecule discovered in 1972 in soil bacteria on Easter Island that is approved by the U.S. Food and Drug Administration for use in cancer therapies and other medical procedures.
"Upon introduction of this small molecule, the two separate inactive fragments of the adenine base editor are glued together and rendered active," Zeng said. "As the body metabolizes the rapamycin, the two fragments disjoin, deactivating the system."
The researchers found some additional benefits to splitting the gene editor in two.
"Compared to an intact editor, our version reduces overall off-target edits by over 70% and increases the accuracy of on-target edits," Zeng said.
In collaboration with Zheng Sun, associate professor in the Department of Molecular and Cellular Biology and in the endocrinology, diabetes and metabolism division of the Department of Medicine at Baylor College of Medicine, researchers targeted the PCSK9 gene, which serves as the blueprint for a protein that helps regulate blood cholesterol levels.
"We hope to see the eventual application of our split genome-editing tool with higher precision to address human health-related questions in a much safer way," said Gao, the Ted N. Law Assistant Professor of Chemical and Biomolecular Engineering.
More information: Hongzhi Zeng et al, A split and inducible adenine base editor for precise in vivo base editing, Nature Communications (2023). DOI: 10.1038/s41467-023-41331-5
Journal information: Nature Communications
Provided by Rice University