This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Structural insights illuminate the arms race between crop plants and fungal pathogens
Many cereal crops, such as wheat and barley, are prey to devastating fungal diseases caused by infection with so-called grass powdery mildews. A key battleground between the plants and the powdery mildews is the interaction between plant immune receptors and pathogen effectors, molecules which are delivered into host cells by pathogens to establish infection.
These effectors and immune receptors are locked in a molecular arms race in which the fungus needs to continuously adapt its effector repertoire to avoid recognition by adapted immune receptors and maintain virulence activity.
However, the structures and functions of these numerous effectors—which can run into the hundreds for individual fungal lineages—remain incompletely characterized.
Now, scientists from Germany, Switzerland and China led by Paul Schulze-Lefert, director at the Max Planck Institute for Plant Breeding Research in Cologne and Jijie Chai who holds positions at Westlake University, Hangzhou and Tsinghua University, both in China, have reported the structures of several powdery mildew effectors from different subfamilies.
These structures show how effectors adopt a common structural scaffold with some local variations that allow them to evade recognition by immune receptors. Their findings are published in the Proceedings of the National Academy of Sciences.
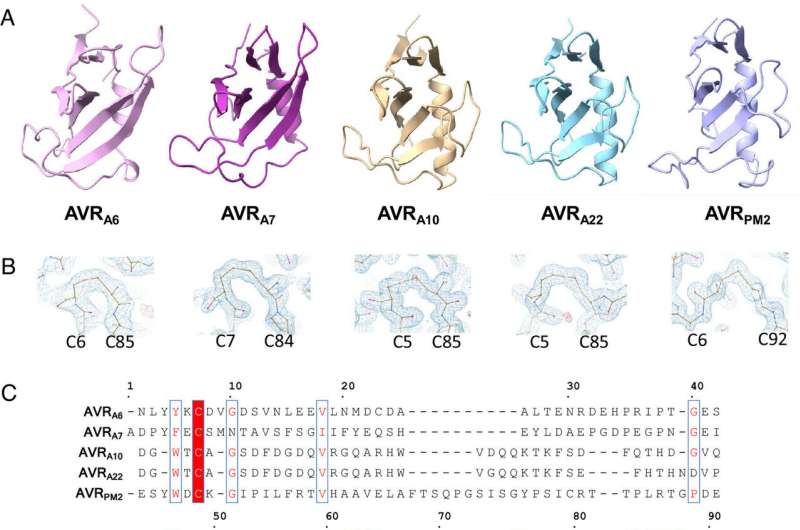
Using X-ray crystallography, a technique that allows scientists to deduce the positions of atoms in a molecule based on electron density, first authors Yu Cao and Florian Kümmel and colleagues acquired structures for five different effectors from two different powdery mildews that infect barley and wheat. Strikingly, although the similarity between the effectors was very low at the level of DNA sequence, they were all found to adopt a common structural fold, known as RALPH after RNase-like proteins associated with haustoria.
Analysis of these structures revealed that they are indeed similar to that of RNase proteins, enzymes that bind to, and break down RNA molecules. Intriguingly, however, these effectors do not possess any RNase activity. Instead, the authors suggest that this common fold RALPH framework may be important for critical processes related to infection, such as assembly into functional effectors and the ability to cross biological membranes. The local structural changes in the RALPH scaffold may explain why the effectors can associate with different host proteins to allow infection.
Armed with an understanding of the structural template of a RALPH effector, the researchers then set out to determine whether they could engineer recognition between immune receptors and effectors in instances where effector divergence had led to immune escape.
Strikingly, they found that six amino acid substitutions were sufficient to turn a sequence-diverged effector into one that was recognized by a specific immune receptor. Analysis of further effector-receptor pairs allowed the authors to conclude that each immune receptor detects largely distinct patches on the surface of its corresponding effector.
"It is one of the eureka moments of science when, in evolution, the molecular arms race between plants and pathogens can be explained by local structural changes within a shared three-dimensional protein architecture," says Paul Schulze-Lefert.
More information: Yu Cao et al, Structural polymorphisms within a common powdery mildew effector scaffold as a driver of coevolution with cereal immune receptors, Proceedings of the National Academy of Sciences (2023). DOI: 10.1073/pnas.2307604120
Journal information: Proceedings of the National Academy of Sciences
Provided by Max Planck Society