This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Scientists create high-quality chromosome-level reference genome of Chelidonichthys spinosus

Chelidonichthys spinosus, commonly known as the spiny red gurnard in the order Scorpaeniformes, is a typical migratory fish that undertakes offshore overwintering migrations in late autumn and early winter, and returns to nearshore areas for spawning in the spring.
It has a snow-white belly, reddish-brown body, and underneath its gills, it possesses a pair of colorful, shimmering green fluorescent "butterfly wings." Below these wings, there are three pairs of appendages resembling ballet dancers feet. In reality, these are six separate pectoral fins that are highly flexible and contain taste buds that aid in foraging.
In order to investigate the evolutionary mechanisms behind the unique morphological features and ecological adaptations of C. spinosus, a research team led by Prof. Zhang Hui from the Institute of Oceanology of the Chinese Academy of Sciences (IOCAS) has conducted deep sequencing of its genome using Illumina short-read, PacBio long-read, and Hi-C techniques. They obtained high-quality chromosome-level reference genome of C. spinosus in the Yangtze River Estuary. The results were published in Scientific Data on July 12.
The results showed that the genome size of C. spinosus is approximately 624.7Mb, with a contig N50 length of about 13.77Mb and a scaffold N50 length of about 28.11Mb. A substantial 99.29% of the initially assembled sequences were successfully anchored to 24 chromosomes, and the genome contained 35.96% repeat sequences and 8,235 non-coding RNAs.
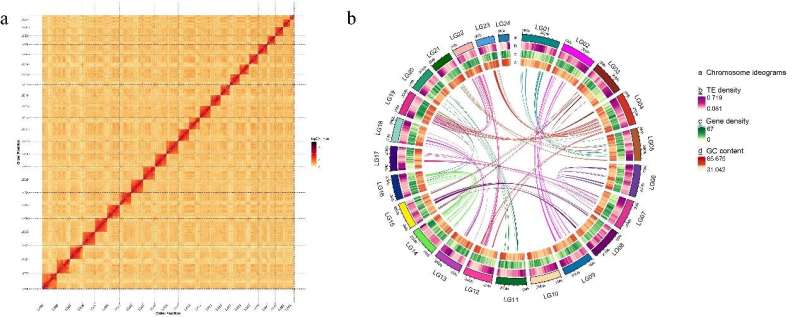
A total of 25,358 protein-coding genes were predicted, of which 24,072 genes (94.93%) were functionally annotated. Co-linearity analysis showed a significant co-linearity between C. spinosus and Cyclopterus lumpus, suggesting a potentially close phylogenetic relationship between the two species.
"The assembled genome sequences can provide valuable information for elucidating the genetic adaptation and potential molecular basis of C. spinosus, which can be used to establish more effective management and conservation strategies for this species," said Wang Yibang, first author of the study.
"These genomic data can also be used for future comparative genomics and phylogenetic studies, which may shed light on the genomic evolution and phylogeny of Scorpaeniformes and other teleost fish and vertebrates," added Prof. Zhang.
More information: Yibang Wang et al, Chromosome genome assembly and annotation of the spiny red gurnard (Chelidonichthys spinosus), Scientific Data (2023). DOI: 10.1038/s41597-023-02357-y
Journal information: Scientific Data
Provided by Chinese Academy of Sciences