This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Pseudo-chromosome–length genome assembly for the deep-sea eel Ilyophis brunneus sheds light on deep-sea adaptation
Recently, the research team led by Dr. Shunping He from the Institute of Deep Sea Science and Engineering, Chinese Academy of Sciences, published their research findings in the online version of Science China Life Sciences.
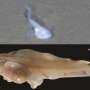
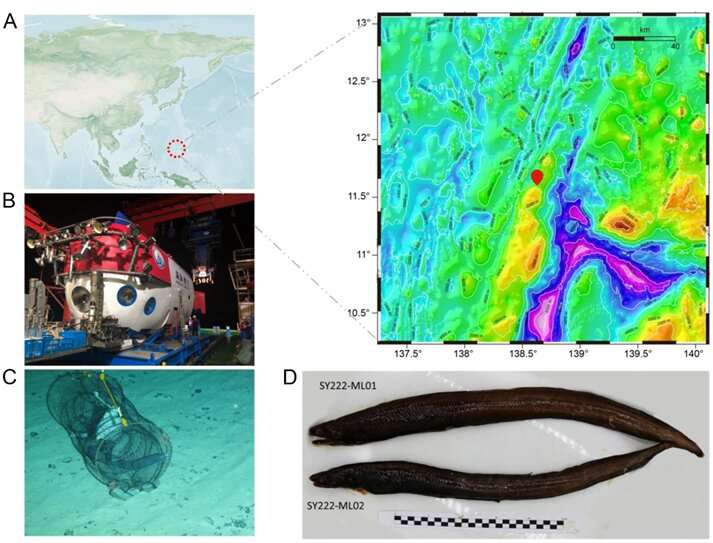
They reported the first high-quality genome of a deep-sea eel (Ilyophis brunneus), which sheds light on the molecular mechanisms of deep-sea adaptation. The deep-sea eel sample was obtained by China's manned submersible, "Shen Hai Yong Shi," at a depth of 3,500 meters in the Mariana Trench. The researchers identified the eel as a muddy arrowtooth eel (MAE) through morphological observation and mitochondrial barcoding analysis.
To investigate the molecular mechanisms of deep-sea adaptation in the eel, the researchers first sequenced and assembled a high-quality genome of the MAE using Illumina high-throughput sequencing, PacBio, and Hi-C technologies. They then conducted phylogenetic and comparative genomic analyses to elucidate its origin and adaptation mechanisms.
The study revealed that several critical genes related to maintaining and regulating the cytoskeleton had undergone specific mutations, such as TUGBCP3 and ITGA genes, which experienced strong positive selection. TUBGCP3 is an essential component of the γ-tubulin complex and plays a crucial role in microtubule nucleation in the centrosome, while ITGA promotes microtubule cytoskeleton stability and regulates cytoskeleton assembly.
The researchers also found that a large number of gene families underwent expansion, positive selection, and rapid evolution, which were related to DNA repair capacity, membrane fluidity, normal transcription and translation processes, and energy metabolism.
In addition, the selection pressure analysis of deep-sea eels, European eels, and other shallow-water fish species revealed that the ω value of deep-sea eels was significantly higher than that of other fish species, suggesting that the deep-sea eel likely experienced functional accelerated evolution in the extreme deep-sea environment. These genetic variations may have enabled the deep-sea eel to evolve its ability to adapt to extreme deep-sea environments.
More information: Jie Chen et al, Pseudo-chromosome—length genome assembly for a deep-sea eel Ilyophis brunneus sheds light on the deep-sea adaptation, Science China Life Sciences (2023). DOI: 10.1007/s11427-022-2251-8
Journal information: Science China Life Sciences
Provided by Science China Press