This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
New DNA identification approach could improve monitoring for chronic diseases
Investigators led by Shana Kelley, Ph.D., the Neena B. Schwartz Professor of Chemistry, Biomedical Engineering, and of Biochemistry and Molecular Genetics, have developed a novel approach for identifying sequences of artificial DNA with differing levels of binding to other small molecules.
The approach, detailed in a study published in Nature Chemistry, could help improve the efficiency of diagnostic monitoring for patients with chronic diseases.
Aptamers are sequences of artificial DNA that selectively bind to other small molecules such as peptides, carbohydrates and foreign pathogens. Aptamers can be used for therapeutic purposes in the same way as monoclonal antibodies, and have been used for pathogen and cancer recognition as well as stem cell markers.
However, generating aptamers with different levels of binding strength has remained a major roadblock in the field, according to Kelley.
"The selection part of it is a bit challenging. You can get a lot of false positives and it's not very tunable. You get reagents out the other end, but it's not very systematic," Kelley said.
In the current study, Kelley's team developed a new workflow to streamline this process that enables efficient and quantitative generation of aptamers with desired binding affinities.
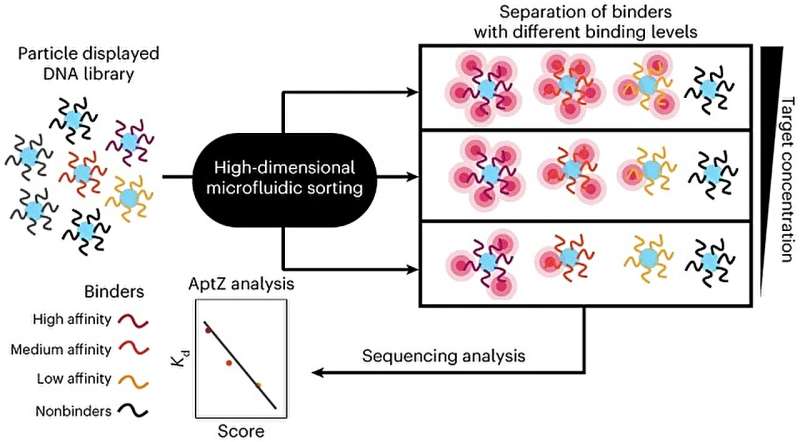
Called Pro-SELEX, the approach uses a molecular analysis technique called microfluidics to isolate a complete set of aptamers with differing levels of binding strength to a particular molecule.
Conventional methods currently rely on repetitive rounds of selection to screen for aptamer candidates. Using Pro-SELEX, the investigators could monitor the binding performance of potential aptamers at different target concentrations in just a single round of selection.
"This turns out to be very handy because sometimes you want very strong binding and sometimes you want weaker binding. There are pros and cons to each, and what comes out of our microfluidic selection are hundreds of these aptamers and we can line up all the sequences and say, 'Well, if we need a really strong binder, we look over here and if we need a less strong binder, we look at this part of the population,'" Kelley said.
According to Kelley, Pro-SELEX could help improve the development of diagnostic sensors and enable more continuous monitoring of biomarkers for patients with chronic diseases such as diabetes, cardiovascular disease or kidney disease.
"If we can develop systems that enable testing more regularly, that is going to help us manage chronic disease much better," Kelley said. "By developing better aptamers that have the right characteristics and give you the right readings, we're hoping we can push towards more continuous monitoring."
More information: Dingran Chang et al, A high-dimensional microfluidic approach for selection of aptamers with programmable binding affinities, Nature Chemistry (2023). DOI: 10.1038/s41557-023-01207-z
Journal information: Nature Chemistry
Provided by Northwestern University